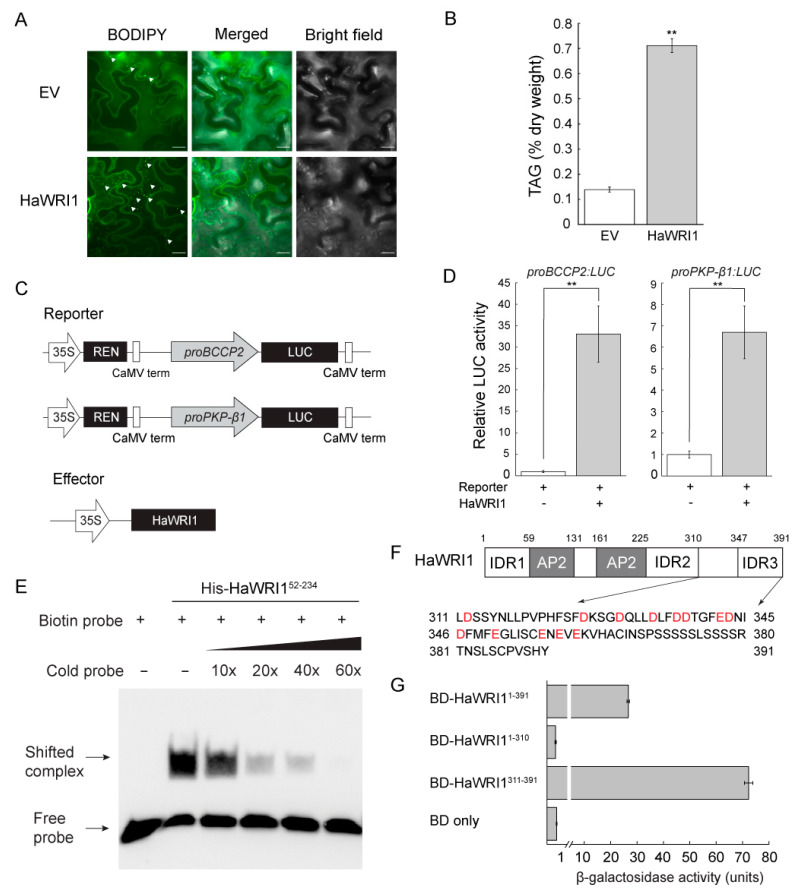
Figure 3.
Functional analysis of HaWRI1. (A) Oil droplets in N. benthamiana leaves transiently expressing HaWRI1. Empty vector (EV) is used as a control. Confocal fluorescence images of N. benthamiana leaf mesophyll tissues displaying oil droplets stained with BODIPY (as indicated by the white arrows) were shown. Scale bar is 20 μM. (B) TAG content in N. benthamiana leaves transiently producing HaWRI1. Results are shown as means ± SE (n = 4). “**” indicates a significant difference (p < 0.01, Student’s t-test) between HaWRI1 and EV. (C) Schematic diagram of constructs used in the dual luciferase assay in N. benthamiana leaves through transient expression. (D) Transactivation activity of HaWRI1 on the promoters of BCCP2 and PKP-β1 (proBCCP2 and proPKP-β1) in N. benthamiana leaves. Results are shown as means ± SE (n = 6). “**” indicates a significant difference (p < 0.01, Student’s t-test) between HaWRI1 and control (reporter without addition of HaWRI1). (E) Binding of the HaWRI1 AP2 domain (amino acids 52–234) to probe containing AW-box in proBCCP2 using EMSA. (F) Schematic diagram of HaWRI1. Acidic amino acid residues in the C-terminal HaWRI1 are highlighted in red color. (G) Transactivation assay of HaWRI1 in yeast cells. HaWRI1 full-length and a series of HaWRI1 truncated variants were subcloned to vector fused with yeast GAL4 DNA binding domain (BD). Transactivation activity was measured via β–galactosidase assay of liquid cultures. Results are shown as means ± SE (n = 3).