Abstract
This article is one of ten reviews selected from the Annual Update in Intensive Care and Emergency Medicine 2022. Other selected articles can be found online at https://www.biomedcentral.com/collections/annualupdate2022. Further information about the Annual Update in Intensive Care and Emergency Medicine is available from https://link.springer.com/bookseries/8901.
Introduction
Research and development of data-driven artificial intelligence (AI), so-called machine learning, in the intensive care unit (ICU) is at an all-time high. Data scientists and physicians are exploring the potential of machine learning in a vast range of domains, including infection management. From both a data science and a medical point of view, infection management in the ICU is an attractive yet challenging research topic: it is a highly complex area where information from several different medical specialties and sources has to be integrated for a single patient. At the same time, there is an urgent need to optimize infection management in the ICU, both for the individual patient – as timely and adequate treatment determines a patient’s survival – and for society – as rising antimicrobial resistance and inadequate treatment results in increased morbidity and mortality and hence increased costs [1]. Evidence-based, data-generated, and automated AI support is expected to help ICU clinicians and antimicrobial stewardship teams take the next step in tackling these problems. Although the main focus of AI research in the ICU has been occurrence of sepsis and its outcome prediction as well as, more recently, almost every aspect of coronavirus disease 2019 (COVID-19), important progress has been made in the infection management field as well [2–4]. In this chapter, we provide an overview of the current stance of AI/machine learning research in different areas of antimicrobial infection management, the barriers that hinder clinical adaptation, and pitfalls for bedside use.
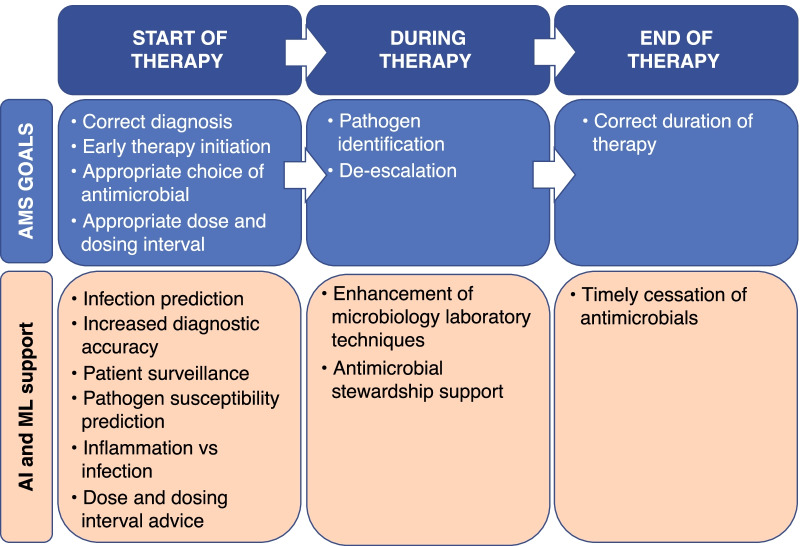
To this end, we have written a narrative review that takes a pragmatic approach using the antimicrobial stewardship cycle as a framework (Fig. 1).
Fig. 1.
The antimicrobial stewardship (AMS) cycle. AI artificial intelligence, ML machine learning
Start of Antimicrobial Therapy
Predicting Infection
A significant number of AI/machine learning models have been developed that try to predict the occurrence of an event in advance, commonly termed ‘forecasting’. Ventilator associated pneumonia (VAP), central-line associated blood stream infections (CLABSI), as well as the risk of colonization/infection with a multidrug resistant pathogen (MDR) are just a few examples for which prediction models have been developed [5–8]. The forecasting of sepsis and/or septic shock has, however, dominated this domain, as illustrated by the no more than 15 retrospective papers and 1 prospective interventional study carried out solely in the ICU that were identified by Fleuren et al. in their recent systematic review [9]. In these and other prediction models, inference of the future risk is made by developing machine learning models on (most often) routinely collected healthcare data (e.g., medical history, clinical parameters, biochemistry results, etc.) from retrospective databases. The rationale behind prediction models is the idea that the clinical course can be altered if the physician is aware of the imminent event. As some predictions can be seen as preventively actionable, clinicians can pre-emptively address known risk factors to try to avert the event from happening. When, for example, the model predicts that a certain risk threshold of CLABSI is exceeded, catheters could be preventively changed or removed to minimize the risk of CLABSI occurrence. Other forecasts however are not preventively actionable from a clinical point of view. Patients at high risk of VAP or sepsis, for example, do not have other known actionable risk factors besides the ones that are already being addressed by standard preventive measures applied today. At first glance, these prediction models will not help in preventing occurrence, but can be used to alert healthcare workers to closely monitor the patient for imminent infection occurrence and consequently facilitate timely initiation of appropriate therapy. Hence, these models could be categorized as early detection or ‘nowcasting’ models. This concept was illustrated in a prospective interventional study by Shimabukuro et al., in which the intervention group was monitored by a machine learning algorithm that alerted the nurse in charge when it identified that the patient was at risk for severe sepsis [10]. The machine learning model was able to make predictions up to 4 h in advance of severe sepsis occurrence. In this study, the mortality and length of stay were significantly lower for patients followed by the machine learning algorithm in combination with the electronic health record (EHR)-based severe sepsis detector compared to patients followed by the latter alone. These findings raise the radical question whether or not we can decrease infection/sepsis related morbidity and mortality by pre-emptively treating selected patients at high risk for infection as predicted by highly accurate machine learning models.
Diagnosing Infection
Increasing Diagnostic Accuracy and Patient Surveillance
Some infections in the ICU are very well described and diagnosis is straightforward. Other infection types have a more subtle clinical course and are defined by different combinations of criteria, making their diagnosis more sensitive to a clinician’s interpretation. In the case of VAP for example, different combinations of clinical symptoms, biochemistry results, radiographic anomalies, and microbiological features can lead to the same diagnosis. When disease criteria are dependent on human evaluation, assistance by AI has the potential to improve interpretation objectivity and hence diagnostic accuracy. Hwang et al. added a machine learning algorithm to human reading of chest X-rays and as a result enhanced the diagnostic performance and accuracy of non-radiologists and radiologists [11]. The implementation of machine learning is not restricted to established diagnostic procedures; new diagnostic approaches are also being combined with machine learning in an attempt to enhance them. Chen et al., for example, explored the possibility of combining electronic nose sensor signals with machine learning for the diagnosis of VAP and attained good accuracy [12].
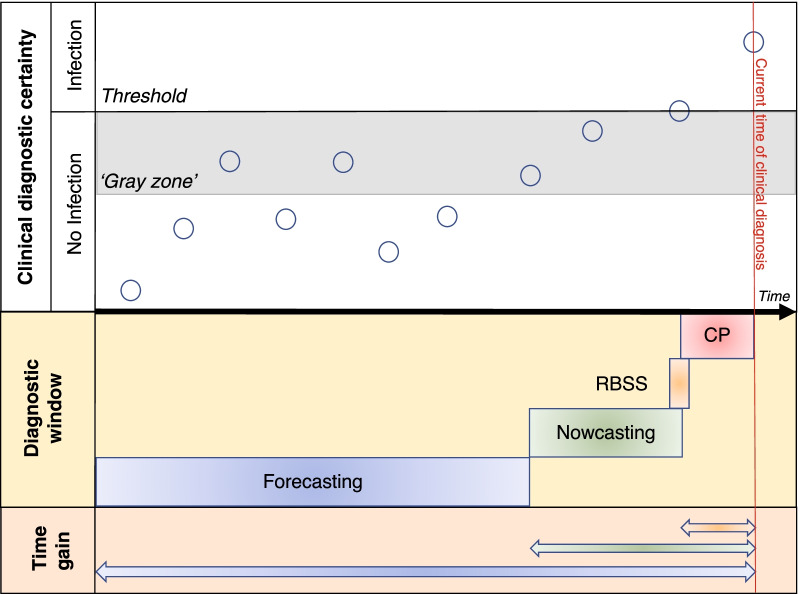
For early infection detection or nowcasting, electronic systems are already being used in clinical practice for automated patient surveillance and early diagnosis of healthcare-associated infections (HAI). Most packages, however, are based on hardcoded rules designed by humans that classify infections as either being present or absent, and hence do not take into account the continuum that is typical of the development of an infection (Fig. 2). A more advanced package is Moni-ICU, whereby first a degree of compatibility (i.e., not compatible, partly compatible, and fully compatible) is expressed between observed/measured patient data and a clinical concept using fuzzy sets (e.g., compatibility between measured blood pressure and heart rate on the one hand, and the concept of shock or drop in blood pressure on the other hand) [13]. Subsequently, combinations of these clinical concepts are being evaluated against higher order concepts (e.g., blood stream infection) by fuzzy rules. Ultimately this leads to the classification of a patient as ‘normal’, ‘borderline infected’ or definitely ‘infected’, and hence early identification of patients in the ‘gray zone’.
Fig. 2.
Continuum of infection development in relation to developing technologies. Current clinical detection of infection is often late in the continuum of infection development (red line). Hard-coded rule-based automatic surveillance systems for early detection only diagnose infection when the clinical threshold of infection has been passed. Fuzzy based surveillance systems are able to identify patients in the preclinical infection zone (“gray zone”), while nowcasting and forecasting models make predictions when infection has not yet been clinically diagnosed. Hence, the time gain to take pre-emptive measures or start appropriate antimicrobial therapy in comparison with current clinical practice can be substantial. O patient state at given time, RBSS rule-based surveillance system, CP current clinical practice
Differentiating Inflammation from Bacterial Infection
From an antimicrobial stewardship point of view, probably the most difficult but also most impactful distinction to make is whether the patient actually has an infection or rather has systemic inflammation without any infection involved. Differentiating between these two disease states requires integration of different types of data, none of which are highly sensitive or specific since abnormal values for these variables is common in both disease states and highly discriminative tests are currently lacking. Lamping et al. however have demonstrated that a machine learning approach, based on random forests using eight routinely available parameters, outperforms currently available biomarkers to discriminate infectious versus non-infectious states in critically ill children [14]. When identification of 100% of sepsis cases was targeted, the model correctly categorized 28% of non-infectious cases. If external validation and clinical trials confirm the validity of the model, it could be associated with a significant reduction in unnecessarily prescribed antimicrobials. A more recent example, where differentiating between inflammation and bacterial infection proved to be burdensome, was the COVID-19 pandemic during which the diagnosis of co-infection of bacterial origin was very difficult to make. This led Rawson et al. to develop machine learning models that support the diagnosis of bacterial infection using only routinely available blood test results [15]. Prospective evaluation of the algorithms is underway, but a preliminary area under the receiver operator curve (AUROC) of 0.96 on 54 patients is encouraging.
Initiating Antimicrobial Therapy
Today, antimicrobials are either prescribed on an empirical basis or complemented with information from surveillance cultures when available. In either case, the causative pathogen is unknown. In addition, the antimicrobial susceptibility of the causative pathogen is only known long after antimicrobial therapy has been initiated. Ideally, rapid diagnostics would lead to the identification of the pathogen and antimicrobial susceptibility directly from clinical samples within approximately 30 min as this would greatly diminish the need for empirical treatment or allow adjustments to the antimicrobial therapy to be made before a second dose is administered, thereby leading to more timely and more appropriate therapy [16]. Also for this domain, researchers have shown that AI/machine learning can play a role.
Enhancing Available Techniques
Machine learning applications have been investigated to enhance currently available phenotypic and genotypic pathogen and resistance identification techniques. Roux-Dalvai et al. for example developed a proteomics library for the 15 most prevalent bacterial species in urinary tract infections (making up 84% of all urinary tract infections) using a liquid chromatography with tandem mass spectrometry technique combined with machine learning, which enables detection of the presence of one of these 15 species within 4 h without the need for bacterial culture [17]. In another study, Feretzakis et al. tested multiple machine learning models that only needed limited information (including source of specimen, presumed site of infection, Gram stain of the pathogen, and previous susceptibility data) to predict susceptibility to a specific antibiotic with 72.6% accuracy in patients admitted to the ICU [18]. But the most promising study from a clinical point of view was by Ho et al. who combined Raman spectroscopy on blood samples and deep learning to develop a base classification model for the 30 most common bacterial and yeast isolate classes in the ICU worldwide [19]. Not only does their method achieve a very high performance in pathogen and resistance identification while only needing ten bacterial cells to function, they also demonstrated that their initial model could be continuously improved with the addition of new Raman spectrums. As the authors state that this technique can process blood, sputum, or urine samples in a few hours without the need for an incubation period, this technique has the potential to greatly diminish the time to pathogen and antimicrobial resistance identification while providing a very high accuracy for certain infections.
Susceptibility Prediction
Although drastically reducing the time to identification, the above mentioned techniques still require sampling and sample processing, and hence will not be able to guide the choice of antimicrobial therapy at the time of initiation. Alternatively, models that can aid in predicting the causative organism and/or antimicrobial resistance at sampling time are also under investigation. Prediction models have been developed, mostly using supervised machine learning, on routinely collected and readily available healthcare data, collected at or before sampling time. A variety of sample types, pathogens of interest, and antimicrobials of concern have already been investigated using this approach with variable degrees of success [20–24]. An advantage of some models is their implementation potential in low and middle income countries. For example, a study performed by Oonsivilai et al. in a Cambodian children’s hospital tested multiple machine learning models to predict the result of the Gram stain and the susceptibility of the pathogen to ampicillin and gentamicin, ceftriaxone or none of the former using only variables derived from clinical and demographic data as well as information regarding their living conditions [24]. Their best performing model had fair predictive performance with an area under the curve (AUC) of 0.71 for the Gram stain result, 0.8 for ceftriaxone susceptibility, 0.74 for ampicillin and gentamicin susceptibility, and 0.85 for resistance to the afore mentioned antimicrobials.
Antimicrobial Dose and Dosing Interval
Pharmacometrics has historically led dose and dosing intervals by means of linear regression, population pharmacokinetic models, and Bayesian forecasting. Developed models are mostly still in the research phase, trying to find their way into the wards as dosing software, but wide implementation in clinical practice is lacking [25, 26]. Introduction of machine learning into pharmacometrics is still in its infancy although the potential of a partnership is increasingly being recognized [27]. At the same time, machine learning research is ongoing to improve antimicrobial dosing, as is illustrated by the vancomycin dose prediction model using XGBoosting developed by Huang et al. [28]. However, more research is needed here as the error is rather high to have potential in clinical practice.
During Antimicrobial Therapy
Machine Learning and the Microbiology Laboratory
By contrast to the pharmacometrics domain, machine learning applications are being explored in all aspects of the microbiology laboratory. For an in-depth review on this topic we refer to the article by Peiffer-Smadja et al. [29]. From microscopic images over spectroscopy data and transcriptomics to gene sequences, no stone is being left unturned. In general, the goal of most of these models is microorganism identification/quantification and evaluation of antimicrobial resistance with the purpose of reducing the turnaround time. As an example, Inglis et al. demonstrated that supervised machine learning could expedite the identification of antimicrobial resistance by using data generated through flow-cytometer-assisted antimicrobial susceptibility testing [30]. Their prototype was able to generate a predictive inhibitory concentration within 3 h of identification of a positive blood culture, where standard methods take approximately 24 h after culture positivity. By combining machine learning with infrared spectroscopy, Lechowicz et al. were able to bring the turnaround time even further down to 30 min, but it should be noted that their machine learning models, based on artificial neural networks, did not achieve perfect classification results [31].
Antimicrobial Stewardship Support
In recent years, a lot of effort has gone into the development and maintenance of hospital tailored antimicrobial stewardship programs. Implementation of these antimicrobial stewardship programs has already had a significant impact on the length of stay and antimicrobial expenditure [1]. One essential element in most of these plans is antimicrobial prescription review and prescriber feedback, where several key parameters for prescribed antimicrobials (e.g., indication, dosage, route of administration, duration) are evaluated. Suggestions are made by the reviewer if adaptations are deemed necessary. Since this is a time-consuming job, computerized systems are often used to help identify patients where a review is warranted. It should however be noted that these clinical decision support systems (CDSS) often have an expert and rule-based knowledge base, which mandates that development and maintenance of these systems to changing guidelines are also time and resource intensive. In addition, resource constraints lead to certain antibiotics of interest being singled out instead of evaluating all prescribed antibiotics. To overcome these shortcomings, Bystritsky et al. tried to develop linear regression and boosted-tree models using routinely available health care data to identify patients that could possibly benefit from prescription review and prescriber feedback [32]. Although the discriminatory power of these retrospectively developed models was only fair and the number of patients that needed to be reviewed by the model to identify one patient who required an intervention was high, the premise that all patients with a prescribed antimicrobial could be evaluated in an automated way could yield a large advantage. Another approach could be to combine the currently available CDSS systems for antimicrobial stewardship with machine learning. Researchers from the Université de Sherbrooke demonstrated that their supervised learning module could identify clinically relevant new rules complementing the rules already in their knowledge base by evaluating past recommendations from clinical pharmacists [33]. Although promising, incorporating new rules, learned through machine learning, into the already available knowledge base and automating rule maintenance remains an important challenge.
End of Antimicrobial Therapy
At present, sometimes empiric antimicrobials are prescribed for patients who do not need it, or they are not stopped in a timely manner. Eickelberg et al. investigated whether machine learning could help identify patients at low risk for bacterial infection and hence suitable for antimicrobial discontinuation [34]. Different machine learning models were investigated, using clinical parameters and characteristics, blood gas and laboratory results, as well as certain administered medications, to evaluate the bacterial infection risk at three time points after initiation of empirical antimicrobial therapy: 24 h, 48 h and 72 h. Interestingly, there was little variation in performance between the 24 h and the 72 h models. The best performing models identified patients with a low risk of bacterial infection with a negative predictive value of more than 93%.
Future Directions
From Research to Clinical Practice
Based on the information provided above, AI and machine learning research are more and more intertwining with every aspect of the antimicrobial stewardship cycle, albeit to varying degrees. However, clinical implementation has only sporadically been realized as most models are still in the design/prototype phase, or have only been tested on internal clinical data [35, 36]. Validation of these models, prior to implementation on external datasets and in clinical trials, will be paramount and adequately designing these trials will be a challenge. Choosing the correct reference standard as a comparator for models for which the ground truth is objective (e.g., prediction of an antimicrobial concentration) is more straightforward than determining this standard for a task where human interpretation and hence subjectivity is involved (e.g., the diagnosis of VAP) [37]. At the same time, a decision will have to be made as to what level of performance we, as clinicians, deem sufficient for a model to be put into clinical practice depending on the given task. Expecting flawlessness from machine learning models is a utopian dream as long as we are not able to incorporate variables that resemble the complete causal pathophysiological process. And even then it is unlikely that models will be flawless, as for even the most objective ground truth, repeated measurements can vary owing to the measurement method used (e.g., the between-run and within-run imprecision of high-performance liquid chromatography with tandem mass spectrometry for antimicrobial concentration determination).
Other aspects that need to be solved are the ethical and legal responsibilities when, for example, clinicians follow off-label dosing suggestions, or follow the advice of a model that later turns out to be flawed. Closely linked to this aspect is the need to educate physicians to critically appraise and evaluate model capabilities and associated studies. As clinicians will be the end-users of these AI systems, education is needed, not only to ensure appropriate usage, but also to empower physicians to identify and report emerging problems.
Finally, all models mentioned in this paper have a standalone design and are focused on one particular aspect of antimicrobial stewardship. Incorporating and managing the variety of engineered models into everyday practice in a meaningful way so that the whole domain of antimicrobial stewardship is supported while not chaining the physician to a screen might pose the greatest challenge of all.
Post-implementation Surveillance of Machine Learning Models
Since deployment of machine learning models into clinical practice is foreseeable in the very near future, governance of these models will be a new task that will have to be taken up by clinicians, at the very least partially. As end-users, clinicians will become the first line of defense to identify circumstances where AI fails to perform reliably. This possibility of failure is not unrealistic, as was recently demonstrated by Finlayson et al. for the proprietary Epic Sepsis prediction model that suffered from a phenomenon called dataset shift [38]. Dataset shift occurs when a machine learning system underperforms after it is deployed because of a mismatch between the data/context it was developed for and the data/context it is deployed in [39]. Translated to clinical practice, this means that any difference or change in patient demographics or delivery of care between the patients the model was developed upon and the actual patients for which the model is asked to give a suggestion, can flaw the suggestion of the model. These differences or changes can be readily identifiable (e.g., evolving antimicrobial resistance epidemiology, or introduction of a new first-line antimicrobial) but can also be very subtle (e.g., behavioral changes of the clinicians induced by the AI system after its implementation, or change of a diagnostic test, which alters the reference values). As clinical practice is changing more rapidly than ever before, keeping models accurate and up to date will be an undertaking in which clinicians will have to take a key role within a medicine-transcendent multidisciplinary team to ensure patient safety. Machine learning solutions such as online learning might prove to be of use in this area as well.
Emerging Research Possibilities
Personalization through the ‘Internet of Things’
The diagnostic process of infection is often triggered by a change in clinical parameters (e.g., body temperature) or laboratory results (e.g., increase in C-reactive protein). For the latter category, increases above a set threshold value as well as trends over time are typically used in clinical practice. For the former category, however, current guidelines use hard thresholds to differentiate a pathological from a normal state, which obviously does not hold true for all patients. For example, fever in adult patients is often defined as a temperature ≥ 38 °C, whereas, depending on the measurement method and age of the patient, the normal population range varies between 35.61 °C and 37.76 °C [40]. Often, older patients do not experience fever while they are having an infection. By integrating information from currently widespread used wearables through the ‘internet of things’, new research opportunities arise to determine –at an individual level– which thresholds should be used to identify significant changes in physiological parameters. Disclosing these baseline physiological characteristics to the physician could help to personalize treatment at a patient level and may also help in the development of machine learning models to do the same. Several companies and healthcare systems have already taken initiatives to enable integration of wearable health technology data into the electronic health record (EHR), but actual clinical impact has not yet been demonstrated [41].
Omics
In addition to AI and machine learning, different types of omics are also being intensively researched in every aspect of healthcare, as it is believed that omics might provide the tools necessary to advance clinical practice toward precision medicine [42]. A difficulty in omics research, however, is the amount of generated data that has to be analyzed and the computational power needed to do so. As AI and machine learning are capable of handling these kinds of issues, combining the two domains might create new insight by integrating information from different omics research fields, as has been illustrated by the ShockOmics research project [43].
Conclusion
AI and machine learning research for antimicrobial stewardship in the ICU is at an all-time high, but to date, implementation into clinical practice has only been sporadic. Internal validation results are promising, so an increase in external validation studies and randomized controlled clinical trials is to be expected in the coming years. Providing the prerequisites to safely validate and implement these models will be necessary for bedside clinical deployment in the near future. Within new beyond-the-borders-of-medicine multidisciplinary teams, bedside clinicians will have an important role in facilitating this process.
Acknowledgements
We would like to thank Jarne Verhaeghe and Femke Ongenae for proofreading the manuscript.
Authors' contributions
All authors contributed to the conception, writing, and revision of the manuscript.
Funding
The work was partially funded by the FWO Junior Research project HEROI2C (Grant nr. G085920N), in which hybrid machine learning for improved infection management in critically ill patients is investigated. Jan J. De Waele is senior clinical investigator and funded by the Research Foundation Flanders (FWO, Ref. 1881020N).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nathwani D, Varghese D, Stephens J, Ansari W, Martin S, Charbonneau C. Value of hospital antimicrobial stewardship programs [ASPs]: a systematic review. Antimicrob Resist Infect Control. 2019;8:35. doi: 10.1186/s13756-019-0471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goh KH, Wang L, Yeow AYK, Poh H, Li K, Yeow JJL, Tan GYH. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat Commun. 2021;12:711. doi: 10.1038/s41467-021-20910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Doorn WPTM, Stassen PM, Borggreve HF, Schalkwijk MJ, Stoffers J, Bekers O, Meex SJR. A comparison of machine learning models versus clinical evaluation for mortality prediction in patients with sepsis. PLoS ONE. 2021;16:e0245157. doi: 10.1371/journal.pone.0245157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamidi ES, Mitsis K, Nikita KS. Artificial intelligence in clinical care amidst COVID-19 pandemic: a systematic review. Comput Struct Biotechnol J. 2021;19:2833–2850. doi: 10.1016/j.csbj.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giang C, Calvert J, Rahmani K, et al. Predicting ventilator-associated pneumonia with machine learning. Medicine (Baltimore). 2021;100:e26246. [DOI] [PMC free article] [PubMed]
- 6.Tacconelli E, Górska A, De Angelis G, et al. Estimating the association between antibiotic exposure and colonization with extended-spectrum β-lactamase-producing gram-negative bacteria using machine learning methods: a multicentre, prospective cohort study. Clin Microbiol Infect. 2020;26:87–94. doi: 10.1016/j.cmi.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Mora-Jiménez I, Tarancón-Rey J, Álvarez-Rodríguez J, Soguero-Ruiz C. Artificial intelligence to get insights of multi-drug resistance risk factors during the first 48 hours from ICU admission. Antibiotics. 2021;10:239. doi: 10.3390/antibiotics10030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parreco JP, Hidalgo AE, Badilla AD, Ilyas O, Rattan R. Predicting central line-associated bloodstream infections and mortality using supervised machine learning. J Crit Care. 2018;45:156–162. doi: 10.1016/j.jcrc.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Fleuren LM, Klausch TLT, Zwager CL, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46:383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimabukuro DW, Barton CW, Feldman MD, Mataraso SJ, Das R. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res. 2017;4:e000234. [DOI] [PMC free article] [PubMed]
- 11.Hwang EJ, Park S, Jin KN, et al. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open. 2019;2:e191095. [DOI] [PMC free article] [PubMed]
- 12.Chen CY, Lin WC, Yang HY. Diagnosis of ventilator-associated pneumonia using electronic nose sensor array signals: solutions to improve the application of machine learning in respiratory research. Respir Res. 2020;21:45. doi: 10.1186/s12931-020-1285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bruin JS, Adlassnig K-P, Blacky A, Koller W. Detecting borderline infection in an automated monitoring system for healthcare-associated infection using fuzzy logic. Artif Intell Med. 2016;69:33–41. doi: 10.1016/j.artmed.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Lamping F, Jack T, Rübsamen N, et al. Development and validation of a diagnostic model for early differentiation of sepsis and non-infectious SIRS in critically ill children—a data-driven approach using machine-learning algorithms. BMC Pediatr. 2018;18:112. doi: 10.1186/s12887-018-1082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawson TM, Hernandez B, Wilson RC, et al. Supervised machine learning to support the diagnosis of bacterial infection in the context of COVID-19. JAC Antimicrob Resist. 2021;3:dlab002. [DOI] [PMC free article] [PubMed]
- 16.Burnham C-AD, Leeds J, Nordmann P, O’Grady J, Patel J. Diagnosing antimicrobial resistance. Nat Rev Microbiol. 2017;15:697–703. [DOI] [PubMed]
- 17.Roux-Dalvai F, Gotti C, Leclercq M, et al. Fast and accurate bacterial species identification in urine specimens using LC-MS/MS mass spectrometry and machine learning. Mol Cell Proteomics. 2019;18:2492–2505. doi: 10.1074/mcp.TIR119.001559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feretzakis G, Loupelis E, Sakagianni A, et al. Using machine learning techniques to aid empirical antibiotic therapy decisions in the intensive care unit of a general hospital in Greece. Antibiotics (Basel) 2020;9:E50. doi: 10.3390/antibiotics9020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho CS, Jean N, Hogan CA, et al. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat Commun. 2019;10:4927. doi: 10.1038/s41467-019-12898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roimi M, Neuberger A, Shrot A, Paul M, Geffen Y, Bar-Lavie Y. Early diagnosis of bloodstream infections in the intensive care unit using machine-learning algorithms. Intensive Care Med. 2020;46:454–462. doi: 10.1007/s00134-019-05876-8. [DOI] [PubMed] [Google Scholar]
- 21.Van Steenkiste T, Ruyssinck J, De Baets L, Decruyenaere J, De Turck F, Ongenae F, Dhaene T. Accurate prediction of blood culture outcome in the intensive care unit using long shortterm memory neural networks. Artif Intell Med. 2019;97:38–43. doi: 10.1016/j.artmed.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Moran E, Robinson E, Green C, Keeling M, Collyer B. Towards personalized guidelines: using machine-learning algorithms to guide antimicrobial selection. J Antimicrob Chemother. 2020;75:2677–2680. doi: 10.1093/jac/dkaa222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman KE, Lessler J, Harris AD, Milstone AM, Tamma PD. A methodological comparison of risk scores versus decision trees for predicting drug-resistant infections: a case study using extended-spectrum beta-lactamase (ESBL) bacteremia. Infect Control Hosp Epidemiol. 2019;40:400–407. doi: 10.1017/ice.2019.17. [DOI] [PubMed] [Google Scholar]
- 24.Oonsivilai M, Mo Y, Luangasanatip N, et al. Using machine learning to guide targeted and locally-tailored empiric antibiotic prescribing in a children’s hospital in Cambodia. Welcome Open Res. 2018;3:131. doi: 10.12688/wellcomeopenres.14847.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roggeveen LF, Fleuren LM, Guo T, et al. Right dose right now: bedside data-driven personalized antibiotic dosing in severe sepsis and septic shock—rationale and design of a multicenter randomized controlled superiority trial. Trials. 2019;20:745. doi: 10.1186/s13063-019-3911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai MG, Cotta MO, Abdul-Aziz MH, Roberts JA. What are the current approaches to optimising antimicrobial dosing in the intensive care unit? Pharmaceutics. 2020;12:638. doi: 10.3390/pharmaceutics12070638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch G, Pfister M, Daunhawer I, Wilbaux M, Wellmann S, Vogt JE. Pharmacometrics and machine learning partner to advance clinical data analysis. Clin Pharmacol Ther. 2020;107:926–933. doi: 10.1002/cpt.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X, Yu Z, Wei X, et al. Prediction of vancomycin dose on high-dimensional data using machine learning techniques. Exp Rev Clin Pharmacol. 2021;14:761–771. doi: 10.1080/17512433.2021.1911642. [DOI] [PubMed] [Google Scholar]
- 29.Peiffer-Smadja N, Dellière S, Rodriguez C, Birgand G, Lescure FX, Fourati S, Ruppé E. Machine learning in the clinical microbiology laboratory: has the time come for routine practice? Clin Microbiol Infect. 2020;26:1300–1309. doi: 10.1016/j.cmi.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Inglis TJJ, Paton TF, Kopczyk MK, Mulroney KT, Carson CFY. Same-day antimicrobial susceptibility test using acoustic-enhanced flow cytometry visualized with supervised machine learning. J Med Microbiol. 2020;69:657–669. doi: 10.1099/jmm.0.001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechowicz L, Urbaniak M, Adamus-Białek W, Kaca W. The use of infrared spectroscopy and artificial neural networks for detection of uropathogenic Escherichia coli strains’ susceptibility to cephalothin. Acta Biochim Pol. 2013;60:713–718. [PubMed] [Google Scholar]
- 32.Bystritsky RJ, Beltran A, Young AT, Wong A, Hu X, Doernberg SB. Machine learning for the prediction of antimicrobial stewardship intervention in hospitalized patients receiving broadspectrum agents. Infect Control Hosp Epidemiol. 2020;41:1022–1027. doi: 10.1017/ice.2020.213. [DOI] [PubMed] [Google Scholar]
- 33.Beaudoin M, Kabanza F, Nault V, Valiquette L. Evaluation of a machine learning capability for a clinical decision support system to enhance antimicrobial stewardship programs. Artif Intell Med. 2016;68:29–36. doi: 10.1016/j.artmed.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Eickelberg G, Sanchez-Pinto LN, Luo Y. Predictive modeling of bacterial infections and antibiotic therapy needs in critically ill adults. J Biomed Inform. 2020;109:103540. [DOI] [PMC free article] [PubMed]
- 35.van de Sande D, van Genderen ME, Huiskens J, Gommers D, van Bommel J. Moving from bytes to bedside: a systematic review on the use of artificial intelligence in the intensive care unit. Intensive Care Med. 2021;47:750–760. doi: 10.1007/s00134-021-06446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleuren LM, Thoral P, Shillan D, et al. Machine learning in intensive care medicine: ready for take-off? Intensive Care Med. 2020;46:1486–1488. doi: 10.1007/s00134-020-06045-y. [DOI] [PubMed] [Google Scholar]
- 37.Chen PHC, Mermel CH, Liu Y. Evaluation of artificial intelligence on a reference standard based on subjective interpretation. Lancet Digit Health. 2021;3:e693–e695. doi: 10.1016/S2589-7500(21)00216-8. [DOI] [PubMed] [Google Scholar]
- 38.Finlayson SG, Subbaswamy A, Singh K, et al. The clinician and dataset shift in artificial intelligence. N Engl J Med. 2021;385:283–286. doi: 10.1056/NEJMc2104626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subbaswamy A, Saria S. From development to deployment: dataset shift, causality, and shift-stable models in health AI. Biostatistics. 2020;21:345–352. doi: 10.1093/biostatistics/kxz041. [DOI] [PubMed] [Google Scholar]
- 40.Geneva II, Cuzzo B, Fazili T, Javaid W. Normal body temperature: a systematic review. Open forum. Infect Dis. 2019;6:ofz032. [DOI] [PMC free article] [PubMed]
- 41.Dinh-Le C, Chuang R, Chokshi S, Mann D. Wearable health technology and electronic health record integration: scoping review and future directions. JMIR Mhealth Uhealth. 2019;7:e12861. [DOI] [PMC free article] [PubMed]
- 42.Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2013;5:73–82. doi: 10.1002/wsbm.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aushev A, Ripoll VR, Vellido A, et al. Feature selection for the accurate prediction of septic and cardiogenic shock ICU mortality in the acute phase. PLoS ONE. 2018;13:e0199089. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.