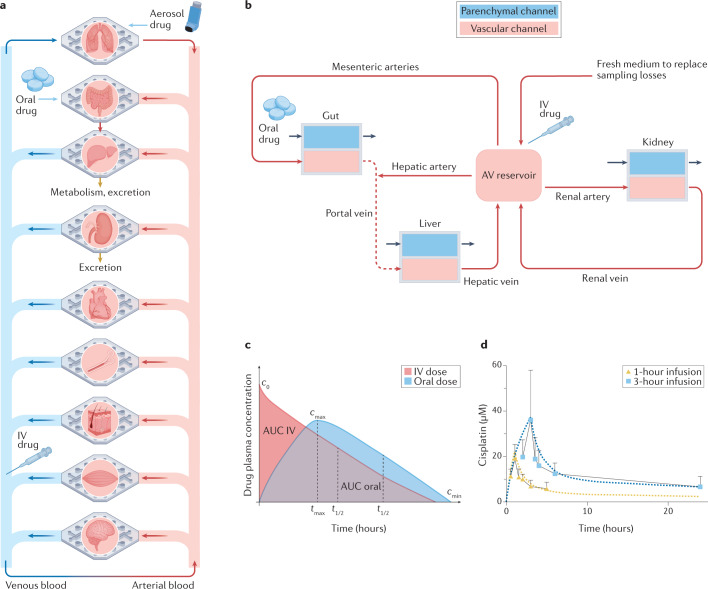
Fig. 3. Modelling drug pharmacokinetics and pharmacodynamics in human body-on-chips.
a | Multi-organ chip systems linked by common flow channels can mimic the physiological linking of organs in our bodies, and hence drug absorption, distribution, metabolism and excretion (ADME) that occurs in the human body as a result of whole body-level physiology can be modelled using this approach. Aerosolized, oral and intravenous (IV) delivery of drugs that occurs in our bodies can be modelled by introducing them into the air space of a lung chip, the lumen of an intestine chip or the vascular channel, respectively; however, IV dosing can be complicated by organ chips immediately downstream from the injection site abnormally experiencing higher doses than other chips due to the lack of mixing that normally occurs in human vasculature. Linked liver and kidney chips can be used to quantify drug metabolism and clearance, respectively, and by linking other relevant chips (for example, a bone marrow chip for myelotoxins), efficacy and potency can be measured as well. b | A schematic diagram showing the fluidic linkages among two-channel intestine, liver and kidney chips corresponding to flow through respective in vivo organ-feeding vessels mimicked by robotic fluid transfers (long arrows indicating flow direction) along with an arteriovenous (AV) reservoir that is fluidically linked to the vascular channels of the organ chips to model blood mixing for more physiologically relevant drug exposures across all chips and to enable experimental sampling analogous to peripheral blood sampling. A common blood substitute is flowed through the vascular channels and the AV reservoir while organ-specific medium is flowed through the parenchymal channel of each chip (small arrows). c | Because the drug levels in effluents of both the vascular and parenchymal channels can be measured over time, pharmacokinetics and pharmacodynamics (PK/PD) parameters — such as area under the curve (AUC), maximum drug concentration in blood (Cmax), and time to reach half-maximal levels (t1/2) — can be determined in vitro using computational physiologically based PK modelling along with scaling approaches. d | This approach has been used to quantitatively predict PK/PD parameters observed in humans in vivo, for example, as shown for cisplatin35, using the body-on-chips linking configuration shown in part b. Squares and triangles indicate PK data obtained from patients in which cisplatin was infused for 1 hour or 3 hours, and dotted lines indicate computational PK predictions generated using data obtained from the human body-on-chips model. The vertical error bars represent the standard deviation. Parts a and b are adapted with permission from ref.148, Annual Reviews. Part c is reprinted with permission from ref.148, Annual Reviews. Part d is reprinted with permission from ref.35, Springer Nature Limited.