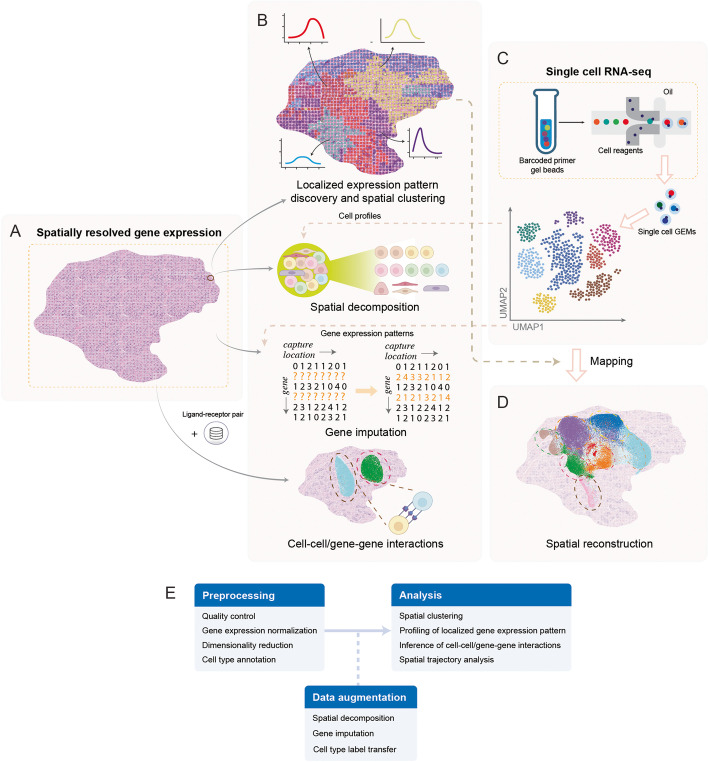
Fig. 1.
Applications of computational approaches in spatial transcriptomics research. A Spatially resolved transcriptomics measures transcriptomes while preserving spatial information. Although spatial transcriptomics data retains spatial information, it is compromised with low cellular resolution and read coverage. B Computational approaches capable of harnessing the complexity of spatial transcriptomics data have been developed for applications of localized gene expression pattern identification, spatial decomposition, gene imputation, and cell-cell interaction. Some of these models leverage gene expression profiles from single-cell RNA-seq (scRNA-seq) data or prior ligand-receptor information from relevant databases to aid spatial transcriptomics research. C Sequencing protocols for scRNA-seq have achieved high-throughput profiling at single-cell resolution, but cellular spatial information is lost during sequencing. Compared to spatial transcriptomics, scRNA-seq is more accessible and can reach cellular resolution. D By leveraging information from the spatial transcriptomics data, spatial location reconstruction could be performed for scRNA-seq data with missing spatial information. In addition, spatial locations could be reconstructed de novo by integrating prior knowledge such as ligand-receptor pair information. E A typical analysis workflow for spatial transcriptomics data. GEMs, Gel beads-in-emulsions; UMAP, uniform manifold approximation and projection