Figure 2.
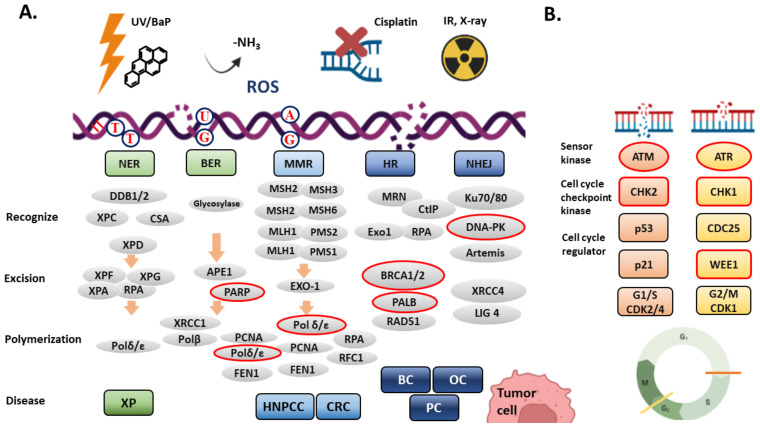
(A) DNA repair systems. Different DNA damages are recognized and repaired through different DNA repair pathways. DNA damages shown here includes ultraviolent light (UV)-induced cyclobutane pyrimidine dimers (TT), benzo[a]pyrene (BaP)-induced bulky DNA adducts, DNA interstrand crosslink, deamination (-NH3, GC to GU pairing), reactive oxygen species (ROS) induced 8-oxoguanine, mismatch DNA pairing (e.g., AG pairing), double-strand breaks (DSBs) induced by ionized radiation, X-ray or cisplatin. Except for direct repair, nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), homologous recombination (HR) and non-homologous end-joining (NHEJ) are shown here. Each DNA repair mechanism comprises three main procedures: recognition, excision, and polymerization. The red circle indicates the defective mutations found in hereditary nonpolyposis colorectal cancer (HNPCC), colorectal cancer (CRC), breast cancer (BC), ovary cancer (OC) and pancreatic cancer (PC). (B) The DNA damage response (DDR). ATM and ATR are activated by DSBs and single-strand breaks. The activated ATM and ATR phosphorylate downstream cell cycle checkpoint kinases, CHK1 and CHK2, which then phosphorylate p53, CDC25, and WEE1. The phosphorylated p53 increases the expression of p21, a potent CDK inhibitor. Phosphorylation of CDC25 (inactive) and WEE1 (active) results in inhibition of CDK activity and leads to cell cycle arrest at G1/S and G2/M transition. Graph created with Biorender.com.