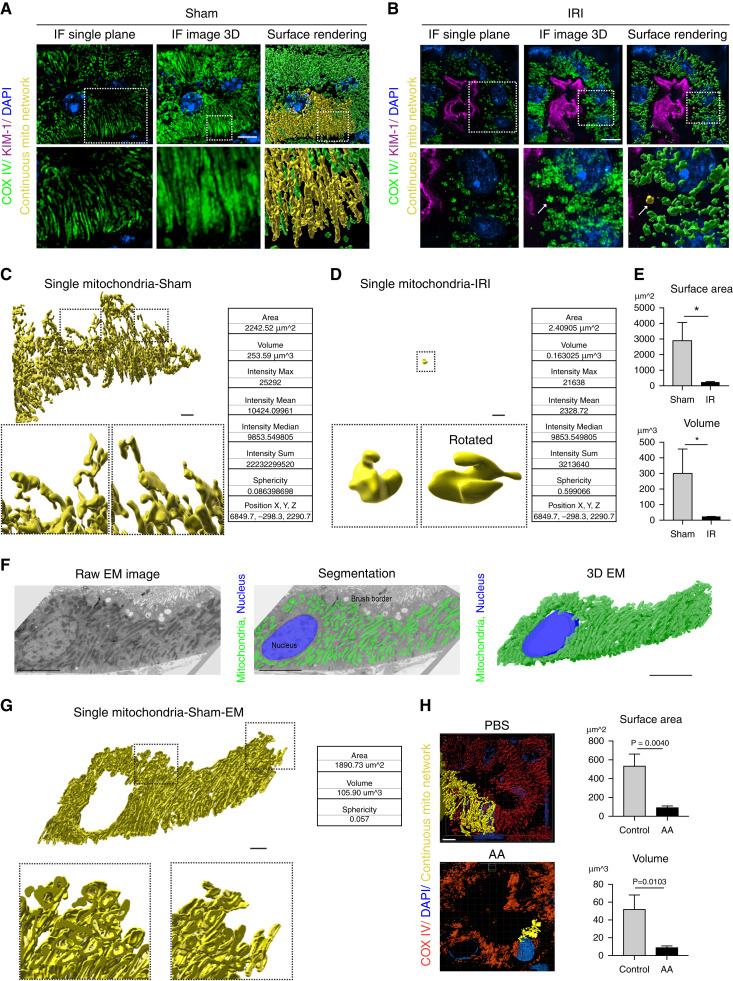
Figure 2.
3D reconstruction reveals kidney tubular cell mitochondria form a large interconnected network. (A, B) Representative images of kidney tubular cell mitochondria stained for COX IV (green) with kidney injury molecule-1 (KIM-1; magenta), imaged by SIM and displayed as a single z-plane (left column), 3D maximum intensity projection (middle), and surface-rendering images (right column) after (A) sham or (B) ischemic-reperfusion injury (IRI) surgery. A continuous mitochondrion is labeled yellow. Arrow indicates fragmented mitochondria. Scale bar, 3 μm. (C, D) Individual mitochondria from the surface-rendering images of (A, B) and sample statistics from the rendered structures. Scale bar, 2 μm. (E) Quantification of total mitochondrial surface area and volume on the basis of surface-rendered images in sham and IRI kidneys. Five tubules per animal were analyzed (n=3). (F) Representative EM image of kidney tubular cells in sham kidney (left). Mitochondria are labeled green in EM image segmentation (middle). 3D structure of mitochondria created by EM serial sections (right). Scale bar, 5 μm. (G) Representative surface rendering of a continuous mitochondrion from the EM segmentation in (F). Scale bar, 2 μm. (H) Representative surface renderings of COX IV–stained (red) mitochondria in kidney tubular cells 6 weeks after PBS or aristolochic acid (AA) injection, and quantification of the total mitochondrial surface area and volume on the basis of surface-rendered images. Five tubules per animal were analyzed (n=3). Scale bar, 4 μm. Data are presented as means±SD. *P<0.05. Dashed insets deliniate the regions shown at higher magnification. (A low-magnification overview of the tubule where the cell was chosen is in Supplemental Figure 1E.) DAPI, 4′,6-diamidino-2-phenylindole; IF, immunofluorescence.