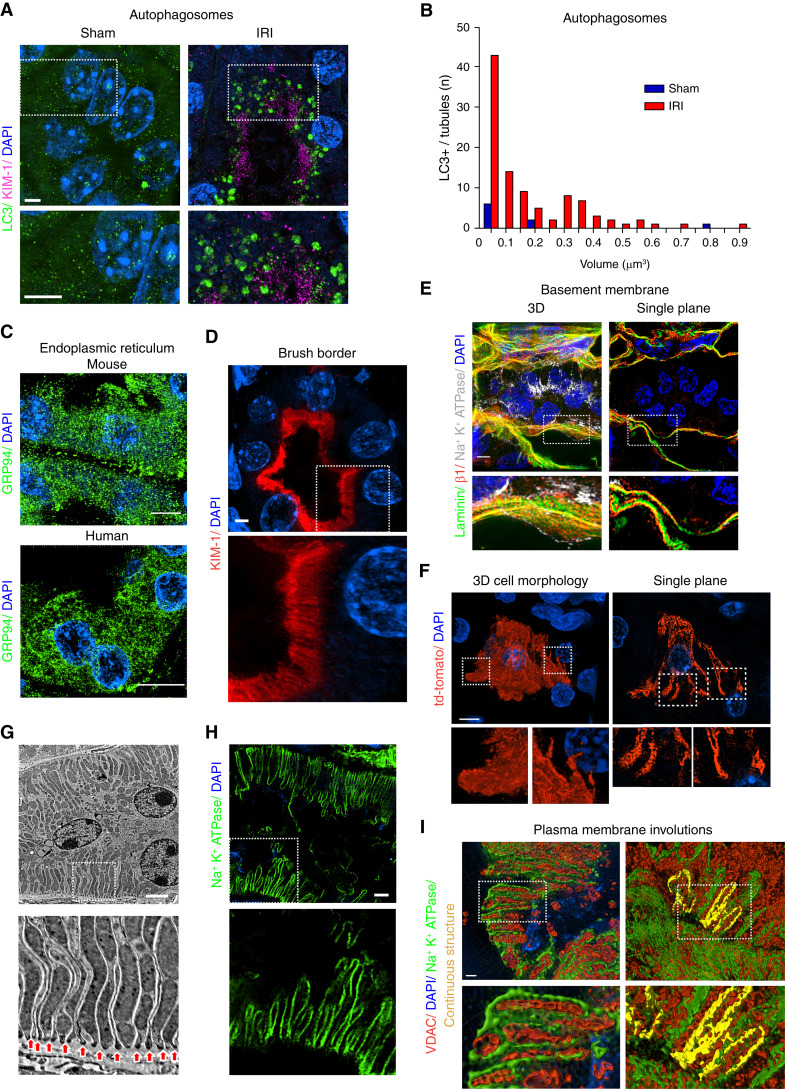
Figure 3.
SIM imaging can resolve multiple organelles and cellular structures in tissue. (A) Representative SIM images of kidney tubules from mice subjected to sham or bilateral IRI surgery stained for LC3 (green). Scale bar, 3 μm. (B) Histogram comparing autophagosome number and size between sham and IRI kidneys. (C) SIM imaging of mouse or human kidney tissue stained for endoplasmic reticulum (ER) using GRP94 (green). (D) Staining of unilateral ureteral obstruction (UUO)–injured mouse kidney tissue for KIM-1 (red), highlighting the brush border. (E) Staining of basement membrane–associated proteins. Uninjured kidneys were stained for laminin (green), integrin β1 (red), and Na+K+ATPase (gray) and imaged by SIM. Maximum intensity projection of a kidney tubule and surrounding basement membrane is shown in left panel. A single z-plane from the same image is shown in right panel. Cutouts show the spatial separation of the proteins. (F) Left panel shows maximum intensity projection of a tdTomato (cytosolic)–positive proximal tubule cell (PTC). Right panels show a single z-plane from a tdTomato-positive tubular cell. (G) EM micrograph of kidney tubular cells highlighting interdigitations of the basolateral plasma membrane (arrows). (H) SIM imaging of tubular cell basolateral membranes. Basolateral plasma membrane is identified by staining for Na+K+ATPase (green). Scale bar, 3 μm. (I) Representative surface renderings of VDAC- (red) and Na+K+ATPase-stained (green) kidneys. Left panel shows high magnification of the basolateral side of a tubule with close association between the Na+K+ATPase and mitochondria. Right panel represents highlighted Na+K+ATPase positive volume showing continuous plasma membrane interdigitation. DAPI, 4′,6-diamidino-2-phenylindole. Dashed insets delineate the regions shown at higher magnification.