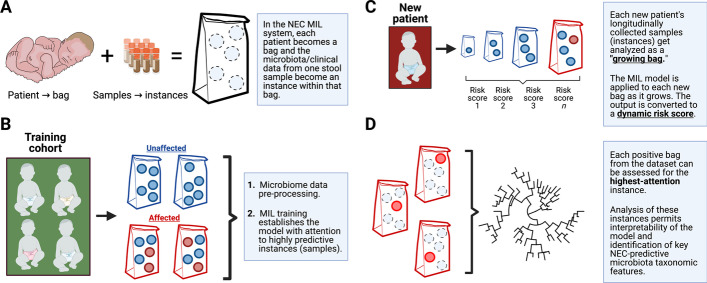
Fig. 3.
Multiple instance learning (MIL) approaches employed in this study. In bag-level MIL, each patient is represented as a bag of multi-feature instances, which are stool samples (the instances) that have been characterized by microbiota states and accompanying basic clinical data (the features). The instances are inherently unlabeled and the goal of the system is to determine a bag label (A). The NEC prediction model was trained on pre-processed microbiota frequency data and basic clinical data from training patient cohorts. This permitted development of a model for bag classification and quantifiable attention to key instances with highest contributions to bag labels (B). Test patients were assessed with the trained model, which was naïve to their data, using a growing bag approach where each new instance generated a new confidence score and attention distribution across all available instances. The changing confidence scores were algorithmically transformed into dynamic risk scores for each test patient (C). Attention scores were also used to identify key, highly informative instances. The feature distributions within these instances were subjected to random forest analysis to identify specific bacterial taxa that drove accurate NEC prediction (D)