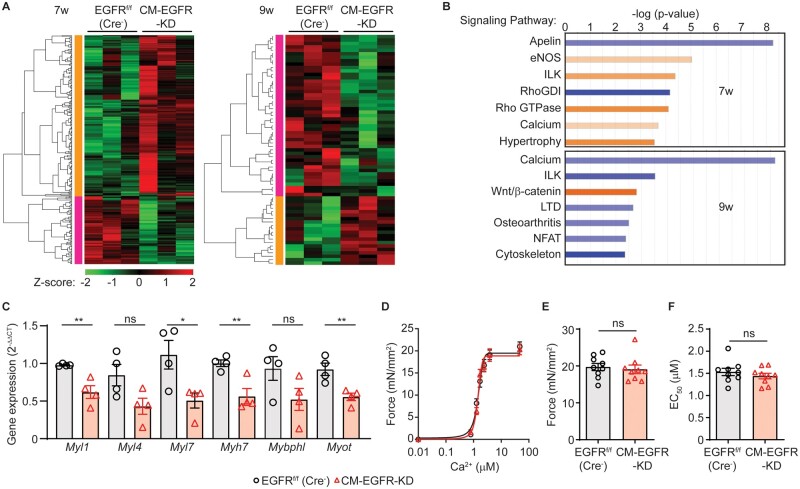
Figure 4.
Cardiomyocyte-specific EGFR downregulation leads to differential temporal effects on cardiac gene expression. (A) Heat maps depicting significant changes in cardiac gene expression in CM-EGFR-KD vs. EGFRf/f (Cre−) hearts (n = 3 each) at 7 (left) or 9 (right) weeks of age. Red and green colours represent upregulated and downregulated genes, respectively. (B) Top signalling pathways altered at 7 (upper panel) and 9 (lower panel) weeks of age in the LV of CM-EGFR-KD vs. EGFRf/f (Cre−) mice as determined by Ingenuity Pathway Analysis (IPA). Blue bars depict downregulation and orange bars upregulation. (C) RT-qPCR was used to measure the expression of Myl1, Myl4, Myl7, Myh7, Mybphl, and Myot in the LV of EGFRf/f (Cre−) vs. CM-EGFR-KD mice (n = 4 each). ns, not significant, *P < 0.05, **P < 0.01, two-tailed unpaired t-test. (D) Mean force as a function of calcium concentration and fitted curves for skinned myocytes isolated from adult EGFRf/f (Cre−) vs. CM-EGFR-KD mice, maximal Ca2+-activated force (Fmax, E) and sensitivity (EC50, F) shown in histograms. Measurements were attained from 3 skinned myocytes per LV isolated from three independent LV samples per genotype.