Fig. 1.
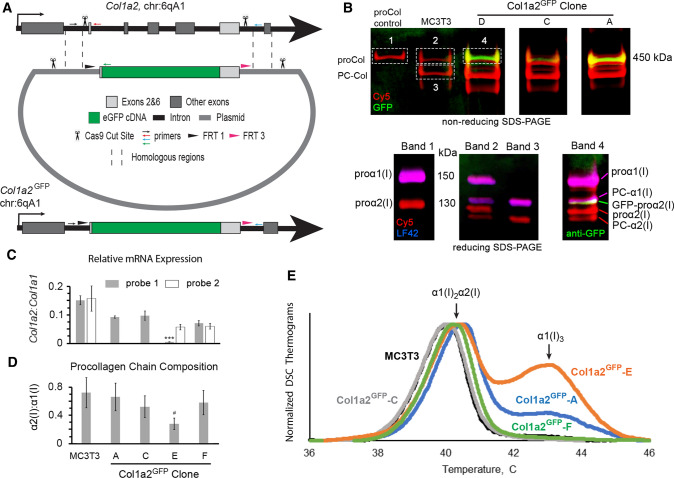
Design of Col1a2GFP MC3T3 murine osteoblasts and procollagen synthesis by different clones. a CRISPR/CAS9 editing scheme at exons 1–7 (gray boxes). Homologous repair of CAS9 cleavage sites in introns 1 and 6 (scissors) based on a donor plasmid with two large homologous arms (dashed lines) replaces exons 2–6 with the following cDNA sequence (5’ → 3’): a Flp recombinase FRT1 site → endogenous intron 1 splice site with full-length exon 2 → eGFP without start and stop codons → full-length exon 6 with endogenous intron 6 splice site → a second, unique Flp recombinase site FRT3. Homologous repair was facilitated by introducing the same CRISPR/CAS9 cleavage sites into the donor plasmid (scissors). The editing was validated by PCR with a common forward primer (small black arrow) and reverse primers for uncut sequence (red arrow), eGFP insertion (green arrow), and additional insertions/deletions (blue arrow). b Top gel shows GFP fluorescence (green) in secreted procollagen molecules labeled with Cy5 (red), which migrate close to procollagen control (proCol) on nonreducing SDS-PAGE. Bottom panel is reducing SDS-PAGE of the main bands cut out from the top gel followed by Western blotting. It shows proα2(I) (Cy5, red) and GFP-proα2(I) chains (red Cy5 and green anti-GFP) integrated into procollagen heterotrimers with proα1(I) (red Cy5 and blue LF42 anti-proα1(I)). PC-Col, PC-α1(I), and PC-α2(I) denote procollagen molecules and chains after cleavage of N-propeptides, respectively. For the first gel, procollagen was denatured at 65 °C to preserve GFP fluorescence. c Relative Col1a2:Col1a1 mRNA ratio (2−ΔCT) in different Col1a2GFP clones measured by qPCR in N = 3 cultures of each clone assuming 100% PCR efficiency. For Col1a2, probe 1 binding at exon 1–2 boundary junction (gray, all cells) and probe 2 binding at exon 34–35 boundary junction (white, MC3T3 and clones E,F) were used. In clone E, probe 1 detected only the Col1a2GFP+ allele, expression of which was ~ 10 times lower than one Col1a2 allele in MC3T3 (***, p < 0.001). d Fluorescence intensity ratio of Cy5-labeled α2(I):α1(I) SDS-PAGE gel bands in pepsin-resistant triple helices purified from cell culture media (N = 6–12), indicating secretion of proα1(I)3 homotrimers by clone E. e Differential scanning calorimetry (DSC) thermograms of pepsin-purified triple helices secreted by wild-type MC3T3 cells and Col1a2GFP clones (0.125 °C/min heating rate in 0.2 M Na-phosphate, 0.5 M glycerol, pH 7.5). The main DSC peak at 40–41 °C represents denaturation of α1(I)2α2(I) triple helices. The second peak at ~ 43 °C is denaturation of α1(I)3 triple helices from proα1(I)3 homotrimers, synthesis of which is caused by deficient expression of Col1a2 [33, 61, 62]. In c, error bars show standard error of the mean (SEM). In d, error bars show estimated assay accuracy (approximately ± 30%), since it was larger than the observed standard deviation. The estimate is based on α2(I):α1(I) ratio variation in control experiments with different cells that produce only α1(I)2α2(I) heterotrimers; # indicates estimated significance. Variation analysis and significance (p value) evaluation are described in Statistical analysis section of Materials and Methods