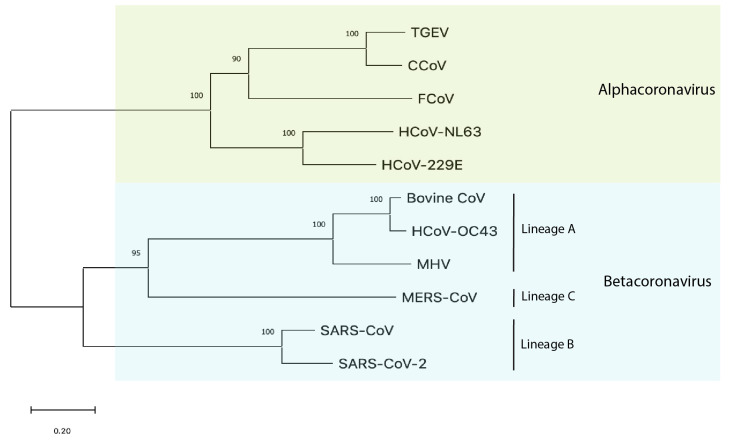
Figure 1.
Phylogenetic tree of the spike proteins of selected coronaviruses. A maximum likelihood phylogenetic tree was constructed using MEGAX (100 bootstraps) from a multiple sequence alignment of spike protein sequences. Spike amino acid sequences were obtained from NCBI GenBank. Accession numbers are: transmissible gastroenteritis virus/TGEV (P07946), severe acute respiratory syndrome coronavirus 2/SARS-CoV-2 (YP_009724390.1), Middle East respiratory syndrome coronavirus/MERS-CoV(AFS88936.1), mouse hepatitis virus/MHV-1 (ACN89742), severe acute respiratory syndrome coronavirus/SARS-CoV (AAT74874.1), feline coronavirus/FCoV-Black (EU186072.1), bovine coronavirus/BCoV (P15777), canine coronavirus/CCoV (AY436637.1), human coronavirus/HCoV-OC43(NC_006213.1), HCoV-229E(NC_002645.1), and HCoV-229E(NC_002645.1).