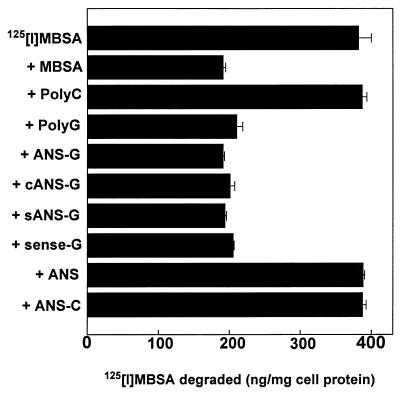
FIG. 2.
Recognition of poly(G) constructs by SCR; competition for degradation of 125I-MBSA. Cells (0.5 × 106 cells/well) were plated in six-well culture vessels and incubated for 18 h at 37°C. Each cell monolayer received 1 ml of RPMI 1640 containing BSA (1 mg/ml) with 2 μg of 125I-MBSA (specific activity: 100 cpm/ng)/ml alone or along with different competitors and was incubated at 37°C. After 5 h of incubation, the medium in each well was processed to determine the amount of 125I-MBSA degraded, as described in Materials and Methods. The molar ratio of 125I-MBSA to different oligonucleotides was 1:20. Unlabeled MBSA was used as a competitor at a concentration of 2 μg/ml. Both poly(G) and poly(C) were 10-nucleotide-long molecules. Results shown are means ± standard errors of three independent determinations.