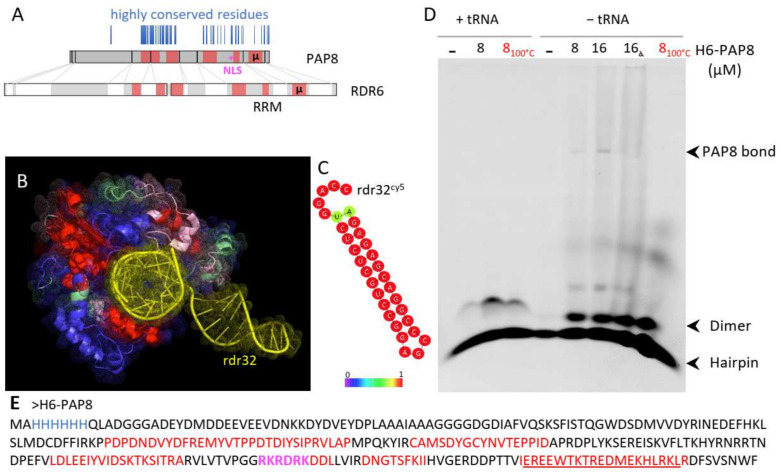
Figure 5.
PAP8 microhomology to RDR6 reveals RNA binding domains. (A) Domain mapping on the amino acid sequence of PAP8 according to the structural homology comparison with RDR6 (pdb:4K50) using T-coffee expresso. Highly conserved residues in the PAP8 orthologous family of proteins are represented as vertical blue lines; µ, amino acid microhomology between PAP8 and RDR6; RRM, RNA recognition motifs in RDR6. (B) Amino acid painting of the three-dimensional fold of RDR6 according to the homology with PAP8: red, good; pink, average; green, bad; blue, absent in PAP8 and yellow, double-stranded RNA sequence named rdr32 hereafter. (C) RNAfold server prediction for the topology of rdr32 RNA sequence used as a cy5-marked probe. (D) RNA electromobility shift assay rEMSA using rdr32cy5 as RNA probe and H6-PAP8 produced in E. coli; 8100 °C, boiled PAP8 protein prior to setting the interaction assay with the probe; 16α, 16 µM of PAP8 + PAP8 antibody (5% of 10,000×). (E) Recombinant H6-PAP8 amino acid sequence; histidine tag in blue; homology regions with RDR6 in red; microhomology underlined and NLS in magenta.