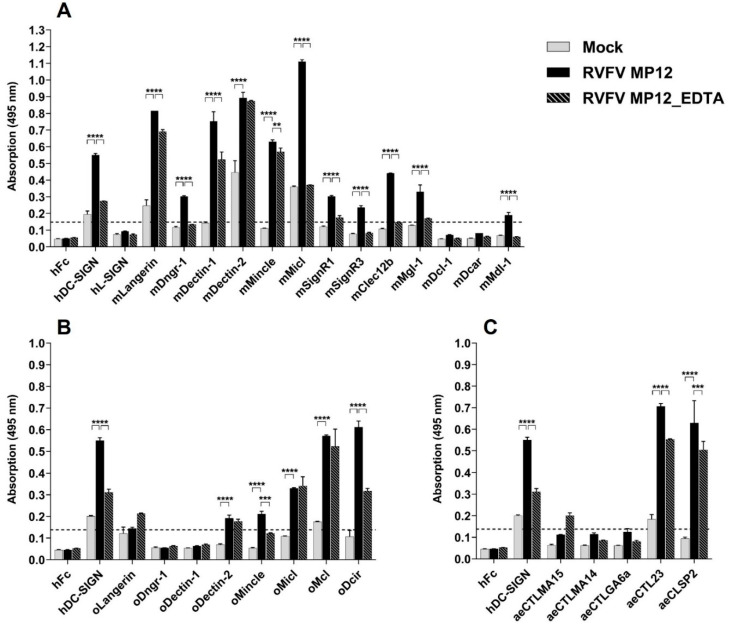
Figure 5.
CLR–hFc fusion proteins across species bind to RVFV MP12: (A) ELISA-based binding study of purified RVFV MP12/Mock with murine CLR–hFc fusion proteins in the presence and absence of EDTA. hDC-SIGN served as positive control [24,45] and hFc-empty as negative control. One representative of n = 5 (EDTA: n = 2) independent experiments is shown; (B) ELISA-based screening of purified RVFV MP12/Mock with ovine CLR–hFc fusion proteins, in the presence and absence of EDTA. hDC-SIGN served as positive control and hFc-empty as negative control. One representative of n = 3 (EDTA: n = 2) independent experiments is shown; (C) ELISA-based screening of purified RVFV MP12/Mock with Aedes aegypti CLR–hFc fusion proteins, in the presence and absence of EDTA. hDC-SIGN served as positive control and hFc-empty as negative control. One representative of n = 3 (EDTA: n = 2) independent experiments is shown; (A–C) to discard possible false positives, the dotted line represents the cutoff defined as the threefold margin of the absorbance value relative to hFc, based on previous screenings with the CLR–hFc fusion protein library [34,46]. Data are depicted as mean + SEM of duplicates. One-way ANOVA with subsequent pairwise Tukey tests were performed to compare the binding of the CLR–hFc fusion proteins above the threshold to mock and the EDTA supplementation; ** p < 0.01, *** p < 0.0005, **** p < 0.0001.