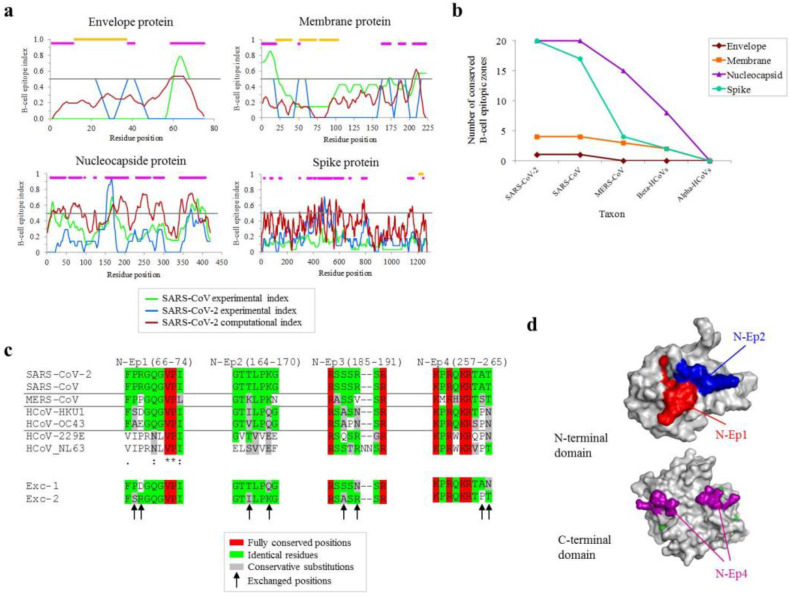
Figure 1.
Conservation of B-cell epitope zones at different taxonomical ranges. (a) Consensus B-cell epitope in the four virion proteins after computational prediction (red line), and experimental validation in SARS-CoV and SARS-CoV-2 applying a window of seven residues. The line indicates the 0.5 threshold applied to select epitopes. Consensus epitopes are in fuchsia. Transmembrane sections are in orange. (b) Taxonomical stepwise conservation of virion betacoronavirus antigens: number of conserved positions globally in the whole alignment (up to 15mers, split into proportional sections if more) extends ≥ 7 positions over 65% similarity and three identical residues, without indels. (c) Alignments of wild-type sequences and proposed consensus peptide sequences for the four most conserved epitopes in the nucleocapside protein. The degree of conservation is color-ranked (see inset legend). Shuffled positions in exchanged epitopes (Exc-1 and Exc-2) are shown (arrow). (d) Structural mapping of conserved epitopes. N-t nucleotide-binding (above) and C-t dimerization (below) domains are shown.