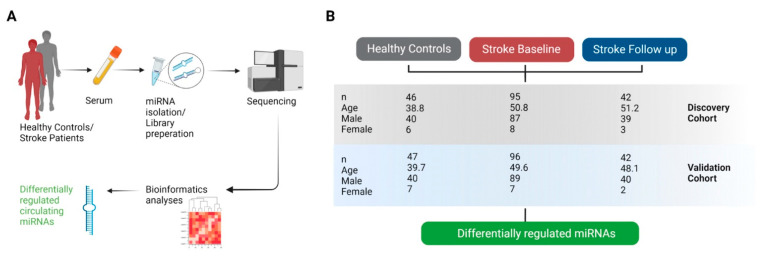
Figure 1.
Study design. (A) Serum samples from stroke patients and healthy controls were collected to isolate circulating miRNAs and generate libraries for RNA-Seq. Multiple bioinformatics tools were utilized for analyses and visualization of sequencing data. (B) Study populations included healthy controls, stroke baseline (BL), and stroke follow-up (FU) patients. Each study population was randomly divided into discovery and validation (replication) cohorts for downstream analyses for the identification of differentially regulated miRNAs.