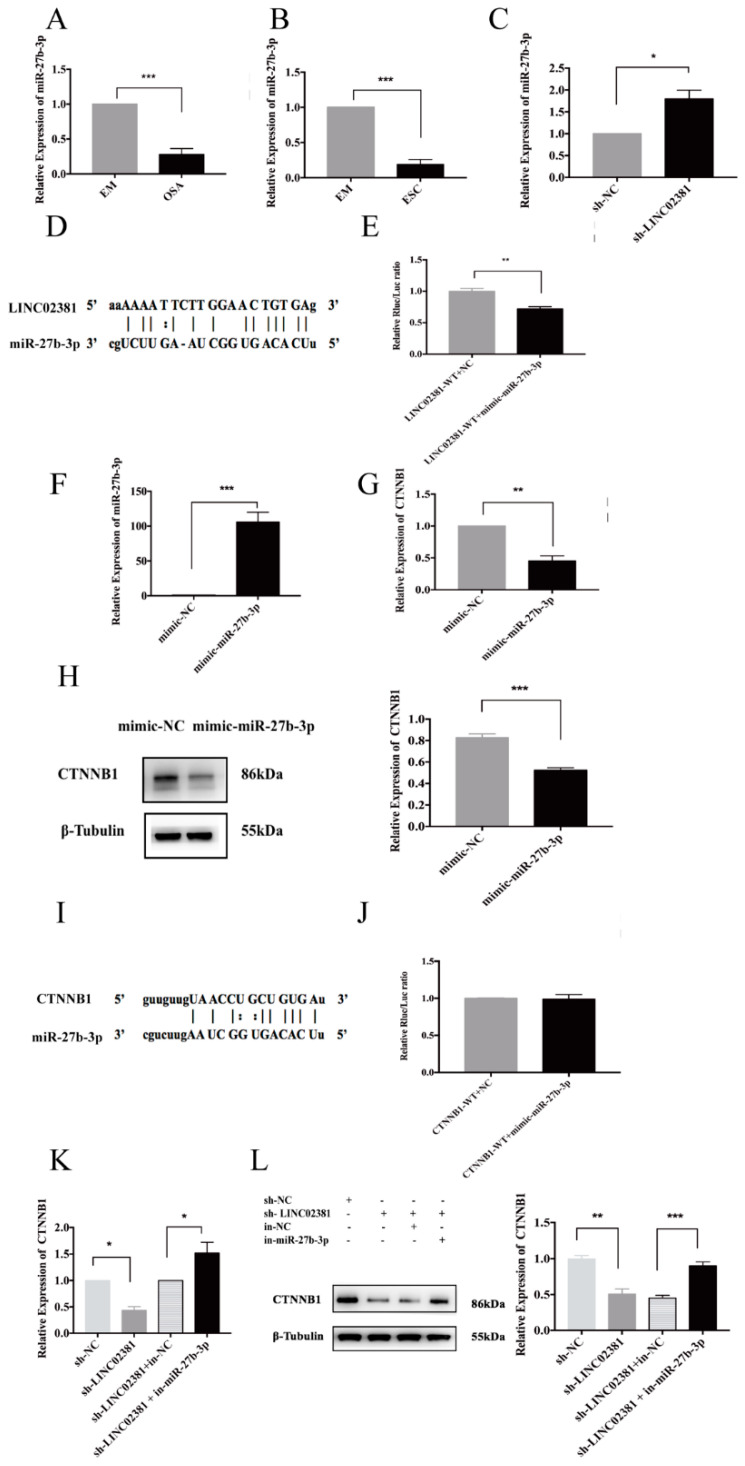
Figure 4.
The LINC02381/miR-27b-3p/CTNNB1 regulatory axis. (A,B) Paired OSA and EM samples and paired ESCs and EMs were collected for qRT-PCR to quantify the RNA expression of miR-27b-3p (n = 15, P < 0.001; n = 12, P < 0.001, separately). (C) ESCs were harvested for qRT-PCR after LINC02381 knockdown (n = 3, P < 0.05). (D) The miRanda software was used to predict the binding relationship between LINC02381 and miR-27b-3p. (E) 293T cells were collected for Dual-Luciferase Reporter Gene Assay after transfection with indicated plasmids (n = 3, P < 0.01). (F,G) ESCs were collected for qRT-PCR after transfection with mimic-miR-27b-3p and mimic-NC. The efficiency of miR-27b-3p overexpression (n = 3, P < 0.001); the mRNA expression of CTNNB1 decreased after overexpression of miR-27b-3p (n = 3, P < 0.01). (H) The protein expression of CTNNB1 in ESCs decreased after miR-27b-3p overexpression (n = 3, P < 0.001). (I) The miRanda software was used to predict the binding relationship between CTNNB1 and miR-27b-3p. (J) 293T cells were collected for Dual-Luciferase Reporter Gene Assay after transfection with indicated plasmids (n = 3, P > 0.05). (K) ESCs were collected for qRT-PCR to quantify the mRNA expression of CTNNB1 after transfection with the indicated plasmids (n = 3, P < 0.05, both). (L) ESCs were cultured for Western blotting to determine the protein expression of CTNNB1 after transfection with the indicated plasmids (n = 3, P < 0.01; n = 3, P < 0.001, separately); * P < 0.05, ** P < 0.01, *** P < 0.001.