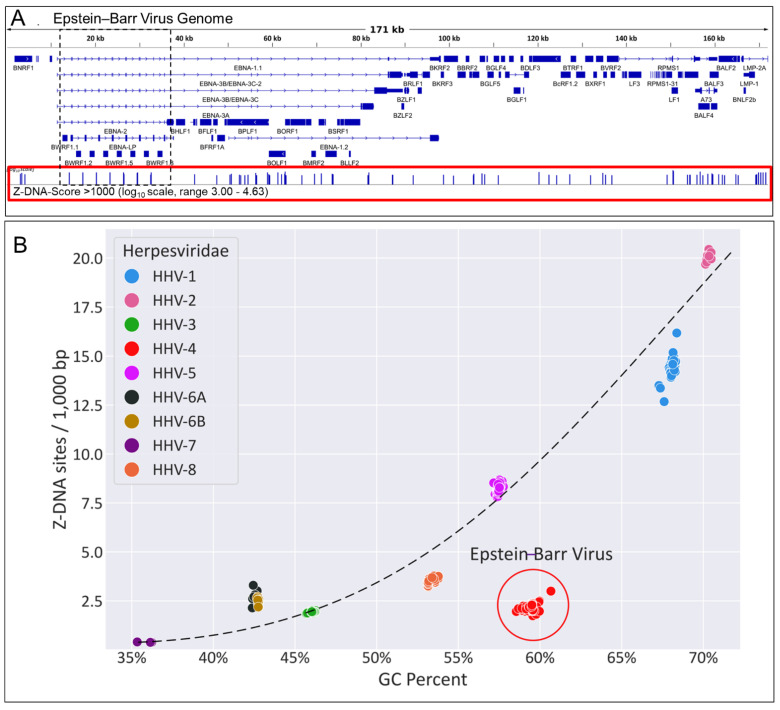
Figure 5.
ZBP1, flipons and disease with evidence of the evolutionary selection against Z-DNA forming regions in the EBV genome. (A). The EBV genome is characterized by strong Z-DNA forming segments in gene promoters. Vertical lines in the red box show the position of sequences with a Z-Score greater than 1000, as determined using the Z-HUNT3 algorithm [2]. Sequences such as these that change their DNA conformation under physiological conditions are called flipons. They can act as switches to turn gene expression “on” or “off” [71]. The region in the dotted box is expanded on panel D of Figure 6 (B). A plot of GC content of different herpes virus (HHV) genome sequences against the number of sequences per 100,000 base pairs with a high propensity to form Z-DNA under physiological conditions (with ZHUNT3 scores >1000). Independent isolates from the various labeled strains are shown. The dashed line represents the expectation that the number of Z-forming sequences increases with GC content. The Epstein virus group of sequences is shifted to the right of this line, consistent with selection against Z-DNA forming sequences in plasma cells.