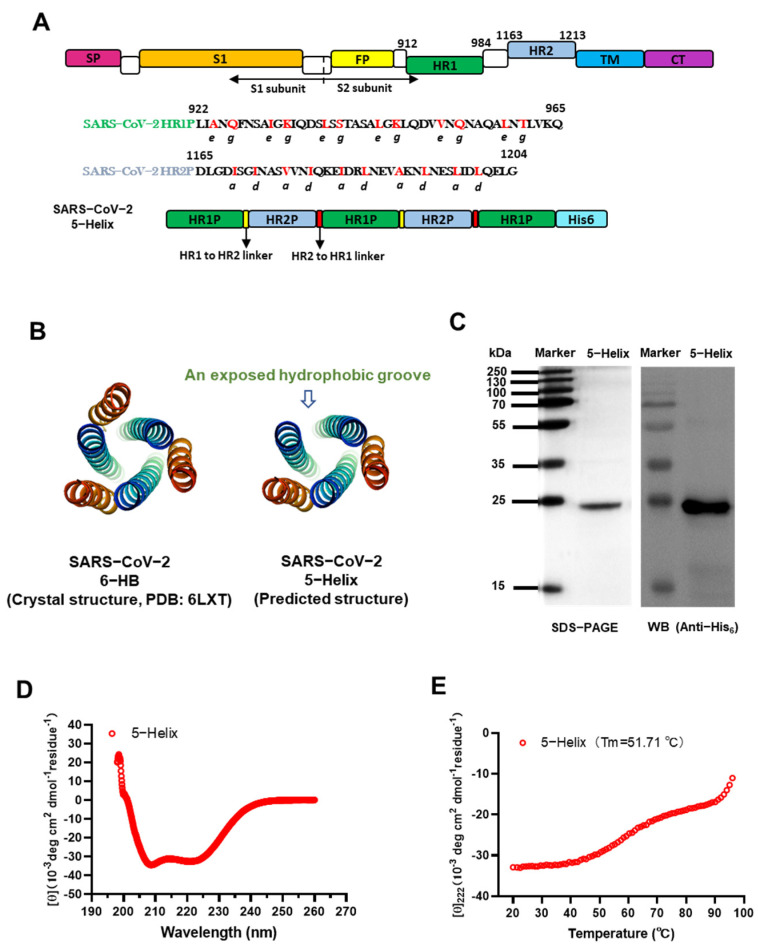
Figure 1.
Design and detection of 5-Helix protein based on HR1 and HR2 sequences of the SARS-CoV-2 S protein S2 subunit. (A) Schematic representation of SARS-CoV-2 S protein and 5-Helix protein. Three copies of HR1 fragment (L922 to Q965, green) and two copies of HR2 fragment (D1165 to G1204, cyan) were sequentially linked with flexible linkers, including the GGSGG linker between HR1 and HR2 linker (yellow) and GSSGG linker between HR2 and HR1 linker (red). SP, signal peptide; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. (B) Predicted structure of 5-Helix and its comparison with the crystal structure of 6-HB formed by the HR1 peptide (cyan) and HR2 peptide (orange) of the SARS-CoV-2 S protein (PDB entry 6LXT, adapted from [7]). (C) Verification of expressed and purified 5-Helix by SDS-PAGE and Western blotting assays. The bacterial lysate was immunoblotted with HRP-labeled anti-His6 antibody; band of the purified 5-Helix was revealed in the SDS-PAGE gel stained with Coomassie blue. (D) The secondary structure of 5-Helix in PBS (pH = 7.4) was examined by CD spectroscopy. Double minima at 208 nm and 222 nm were revealed. (E) Thermal unfolding of 5-Helix. The melting curve and its melting temperature (Tm) are shown.