Fig. 3 |. Engineering biosensor-dependent containment circuits and multiplexing for AND logic gate growth in vitro.
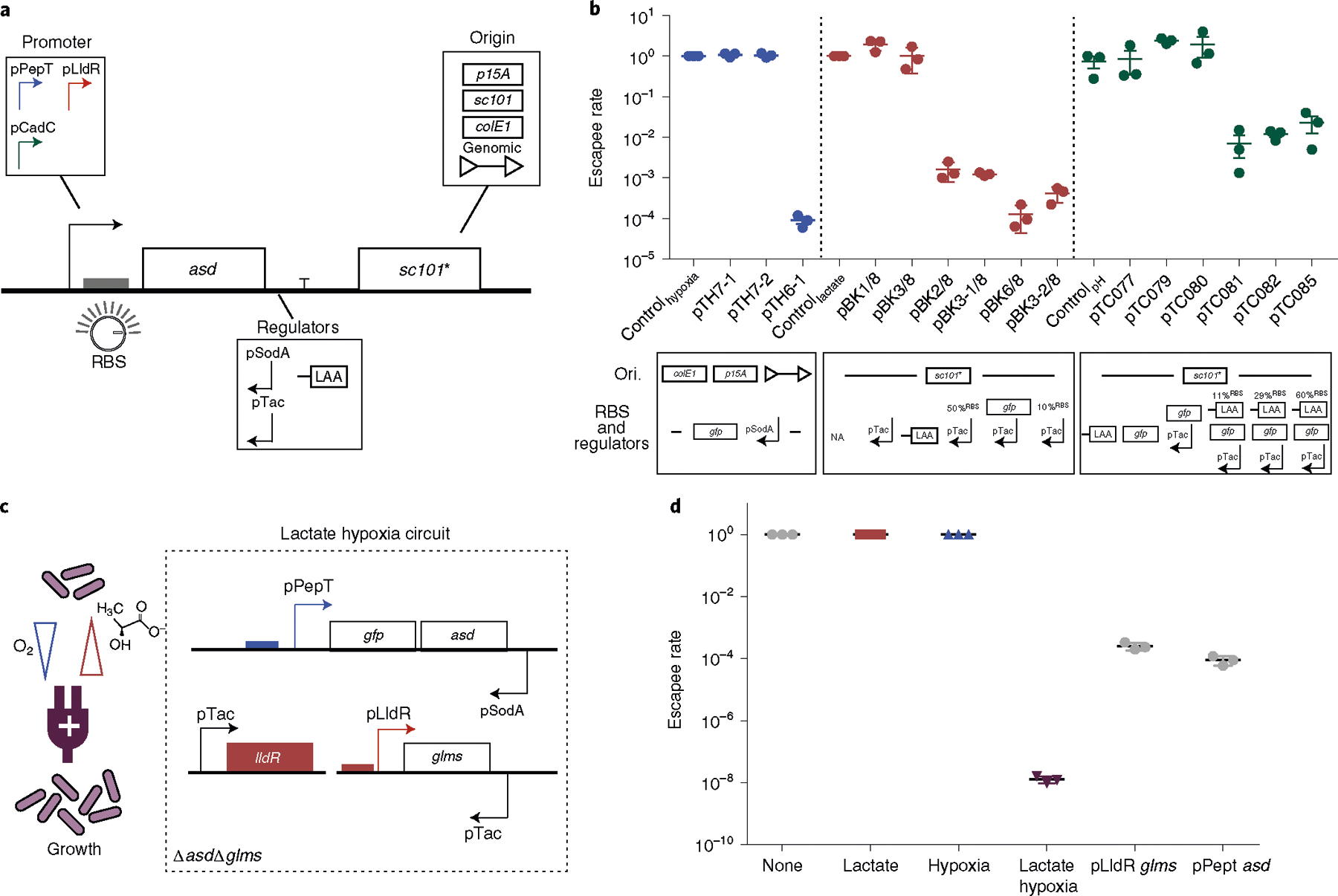
a, Design of a modular containment circuit that includes a biosensor promoter (pPepT, pLldR or pCadC) driving an essential gene (asd). To tune the sensitivity and reduce noise, additional regulators such as antisense promoters (pTac or pSodA), origin of replication (colE1, p15A, sc101* or genome integration), RBS or protein degradation tag (LAA) were used. b, Top: characterization of biocontainment variants on the basis of the escapee rate, defined as the ratio between colonies grown in non-permissive (normoxic, 0 mM lactate and pH 7) and permissive (hypoxic, 10 mM lactate and pH 6) conditions. All variants were cultured for 12–16 h with a starting density of 107 c.f.u. per ml and plated on LB agar plates with added supplements, after which colonies were counted the next day. n = 3 biological replicates. Data are mean ± s.e.m. All variants drove an essential gene along with additional genetic parts such as anti-sense, tuned RBS strength or a degradation tag (LAA). Blue, red and green indicate the hypoxia-, lactate- and pH-driven containment circuit, respectively (Supplementary Tables 2 and 4). Bottom: design of the circuit variants specifying changes in origin of replication (Ori.), RBS and regulators for each construct. NA indicates no modifications in RBS and regulators. c, Schematic of the lactate–hypoxia AND logic gate circuit, which consists of both the hypoxia promoter pPepT driving an essential gene (asd) and the lactate biosensor pLldR driving a second essential gene (glms). d, The escapee rates of the lactate–hypoxia AND logic gate circuit. The engineered bacteria were cultured under the four different conditions (none, 10 mM lactate supplemented, hypoxic conditions and a combination of 10 mM lactate with hypoxic conditions). The escapee rate was calculated as the ratio of non-permissive (no inducers, lactate only or hypoxic only) to permissive (both lactate and hypoxia) conditions. Two single circuits were grown in either lactate or hypoxic conditions. n = 3 biological replicates. Data are mean ± s.e.m. Samples were grown for 16 h and then plated on LB agar supplemented with DAP and d-glucosamine. Colonies were counted after incubating at 37 °C overnight.
