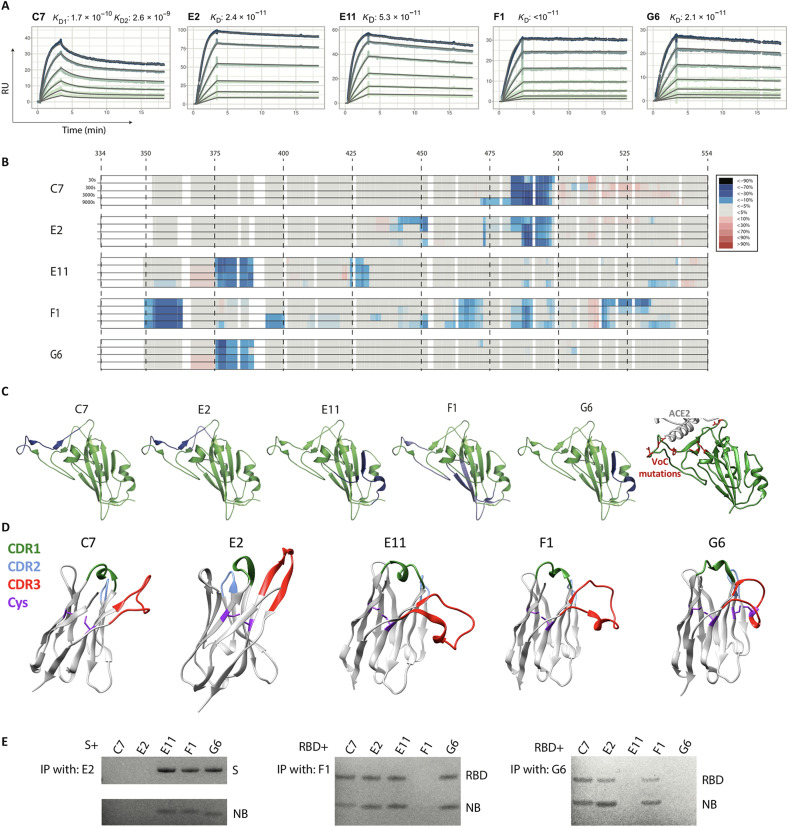
Fig. 5. RBD-targeting affinities and epitope mapping.
Five RBD-targeting nanobodies were selected for affinity characterization by SPR and for epitope mapping by hydrogen/deuterium exchange coupled to mass spectrometry (HDX-MS). (A) Picomolar affinities (170 to <10 pM) for all tested nanobodies. RU, resonance units; KD, dissociation constant. (B) HDX-MS signal across the RBD sequence, which is mapped onto the RBD structure in (C), together revealing three distinct epitope classes. (C) Right-most: The positions of all RBD mutations occurring in variants Alpha, Beta, Gamma, Delta, Kappa, Epsilon, Eta, Iota, and Lambda are shown in red. VoC, variant of concern. (D) AlphaFoldv2 (31, 32) predictions of the structure for these five nanobodies, highlighting the CDRs, and the cysteine pairs. Unlike the others, G6 shows an additional predicted disulfide bond between cysteines in the CDR3 and in FR2. (E) Immunoprecipitation (IP) competition analysis supporting the three epitope classes. C7 and E2, the two most potent neutralizers of SARS-CoV-2, target an epitope at the ACE2 interface, which explains both their potency and their lack of cross-reactivity. Nanobody (NB) G6 targets an epitope that is well conserved across the founder virus of SARS-CoV-2, the Beta variant, and SARS-CoV-1 (where there is only a single substitution relative to SARS-CoV-2), explaining its cross-reactivity, with E11 having a similar epitope. F1 has by far the largest epitope, which potentially explains the very low dissociation rate.