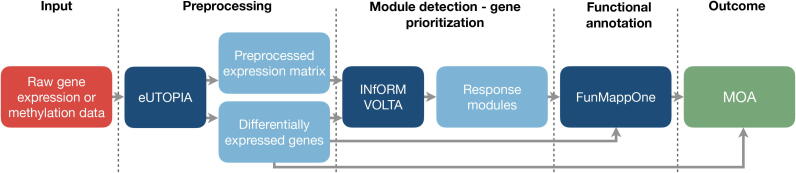
Fig. 2.
Nextcast pipeline for the characterisation of the MOA of a compound. Raw omics data is preprocessed with eUTOPIA. The output of the tool includes a matrix with normalised (and batch corrected) expression values and a list of differentially expressed genes. This data can be fed to INfORM to identify a set of responsive gene modules. VOLTA can be further used to analyse networks built with INfORM. Alternatively, differentially expressed genes can be directly provided as the input for the FunMappOne tool to perform enrichment analysis and identify the underlying biological processes. The result is a list of regulated genes and corresponding enriched pathways or regulated genes in co-expressed modules and their corresponding pathways. The red box represents the input for the pipeline while the green box describes the outcome of the pipeline. The dark blue boxes correspond to the individual Nextcast components of the “Analytics” category, and the light blue boxes indicate the intermediate outputs/inputs.