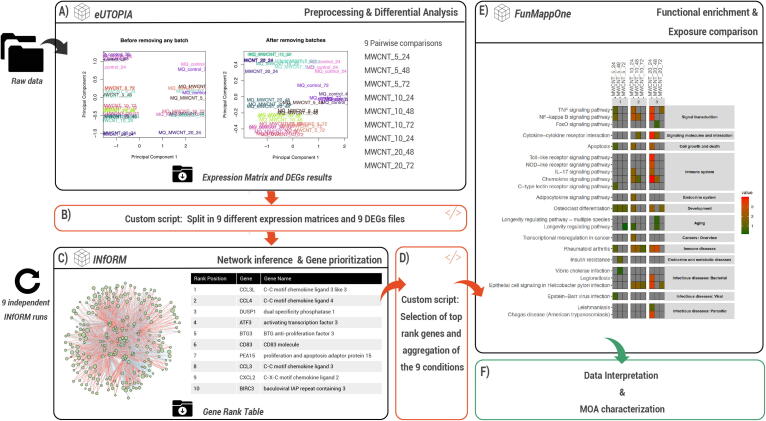
Fig. 7.
Example application of the characterisation of the MWCNT MOA employing INfORM. (A) eUTOPIA was used to preprocess input raw data and to perform differential analysis. The normalised expression matrix, as well as the lists of differentially expressed genes, were exported. (B) A custom script was used to select the most frequently deregulated 1,000 genes across the exposures and to produce inputs for INfORM. (C) INfORM was used to infer the gene co-expression networks and to rank the genes according to their topological properties. (D) The first 200 positions of each list were selected and combined in a format compatible with the FunMappOne input. (E) FunMappOne was used to perform enrichment analysis of the KEGG human pathways. (F) The output was interpreted for MOA characterisation of MWCNT exposures at different doses and time points.