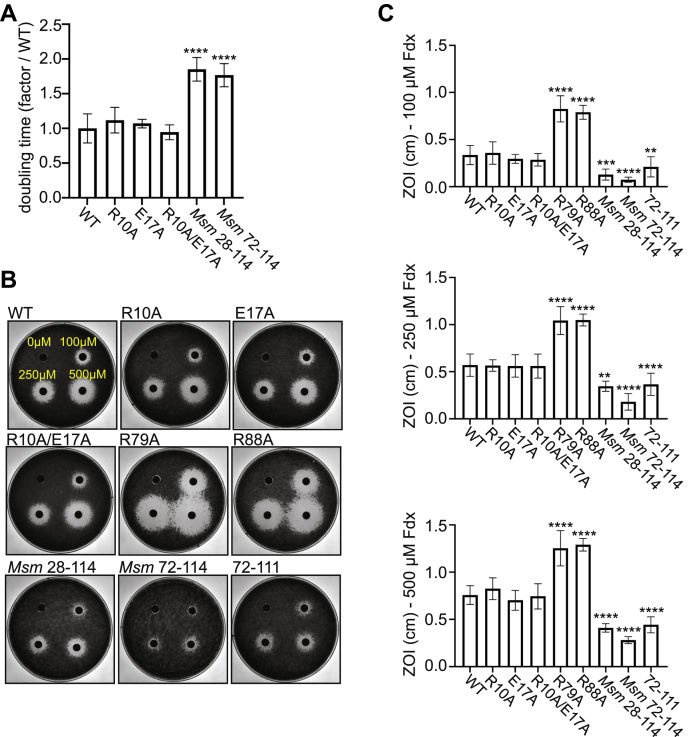
Figure 2.
Multiple RbpA domains impact Fdx activity in vivo.A, ratio of the doubling times of M. smegmatis strains expressing RbpAMtbR10A, RbpAMtbE17A, RbpAMtbR10A/E17A, RbpAMsm28–114, or RbpAMsm72–114 as compared to the average doubling time for the strain expressing RbpAMtbWT. The mean ± SD from at least two independent experiments with three replicates per experiment. B, zones of inhibition (ZOI) by Fdx on bacterial lawns of M. smegmatis expressing RbpAMtbWT, RbpAMtbR10A, RbpAMtbE17A, RbpAMtbR10A/E17A, RbpAMtbR79A, RbpAMtbR88A, RbpAMsm28–114, RbpAMsm72–114, or RbpAMtb72–111 as the only copy of rbpA. C, mean radii of ZOI ± SD from at least two experiments with at least three replicates at 100 μM, 250 μM, and 500 μM Fdx is plotted. For A and C, statistical significance of differences was analyzed by ANOVA and Tukey’s multiple comparison test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. All comparisons to RbpAMtbWT were included in the analysis, but only statistically significant comparisons are indicated in the figure. Fdx, fidaxomicin; RbpA, RNA polymerase binding protein A; RNAP, RNA polymerase.