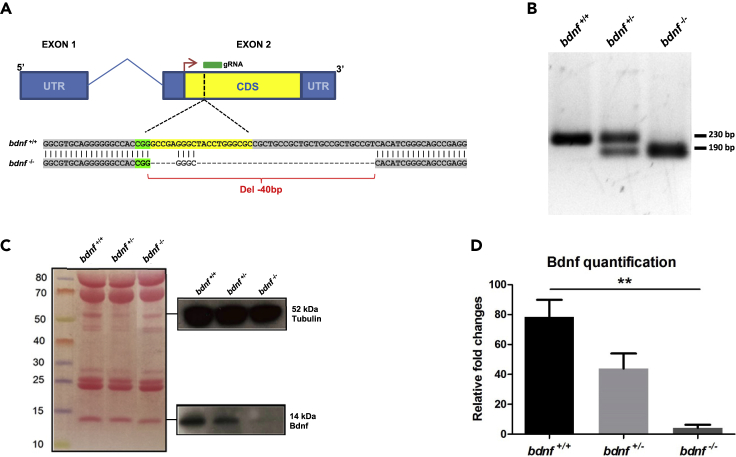
Figure 1.
Generation of bdnf−/− zebrafish line
(A) The bdnf gene structure in Danio rerio is composed of two exons (blue box) and one intron (blue line), with the mRNA coding sequence (CDS) (yellow box) fully present in the exon 2. gRNA designed to target the CDS binds a genomic region immediately after the start codon (red arrow) and determines a 40 bp deletion.
(B) Genomic screening by PCR shows a single band of 230 bp in bdnf+/+ fish, one band of 230 bp and one of 190 bp in the bdnf+/-, and a single 190 bp band in the bdnf−/−.
(C and D) Western Blot analysis and (D) Bdnf quantification show that Bdnf protein is reduced by approximately 50% in bdnf+/- and almost completely in bdnf−/− in respect to bdnf+/+. Tubulin antibody was used as loading control. Densitometric analysis of Bdnf protein from three biological replicates is normalized with respect to Tubulin.
Mean values ±SEM; Dunn’s Multiple Comparison Test: ∗∗p < 0.01.