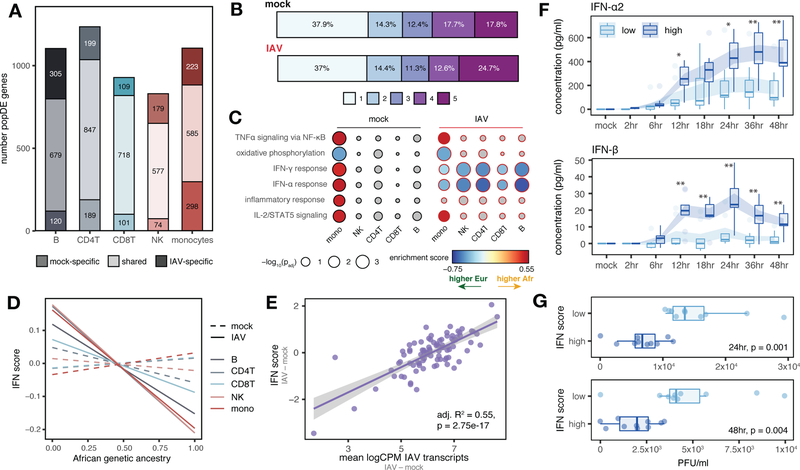
Fig. 2.
Genetic ancestry influences the immune response to IAV infection. (A) Number of shared and condition-specific popDE genes. (B) Cell type sharing of popDE effects (1 = detected in a single cell type, 5 = detected across all cell types). (C) GO enrichments for popDE effects in the mock- and IAV-infected conditions. Colored circles represent pathways with FDR < 0.10. IFN pathways are among the most divergent between European and African-ancestry individuals in monocytes, with 26% (42 out of 163) of all IFN genes tested classified as popDE after infection. (D) Correlation between African genetic ancestry proportion and IFN score in mock (dotted lines) and IAV-infected conditions (solid lines). (E) IAV transcript levels are associated with IFN response in PBMCs. (F) Secreted IFN-α2 and IFN-β levels in low versus high IFN responders over a 48 hour time course. Shaded area represents the mean ± SE. *p < 0.02, **p < 0.009 (Mann-Whitney U tests). (G) Viral titers (plaque-forming units, PFU/ml) detected in supernatant 24 and 48 hpi. In (D) and (E), p-values and best-fit slopes were obtained from linear regression models.