Figure 7. Phylogenetic analysis of the amino acid sequences of ATE1, PHD, HIF1α, pVHL, and Ubr among different species.
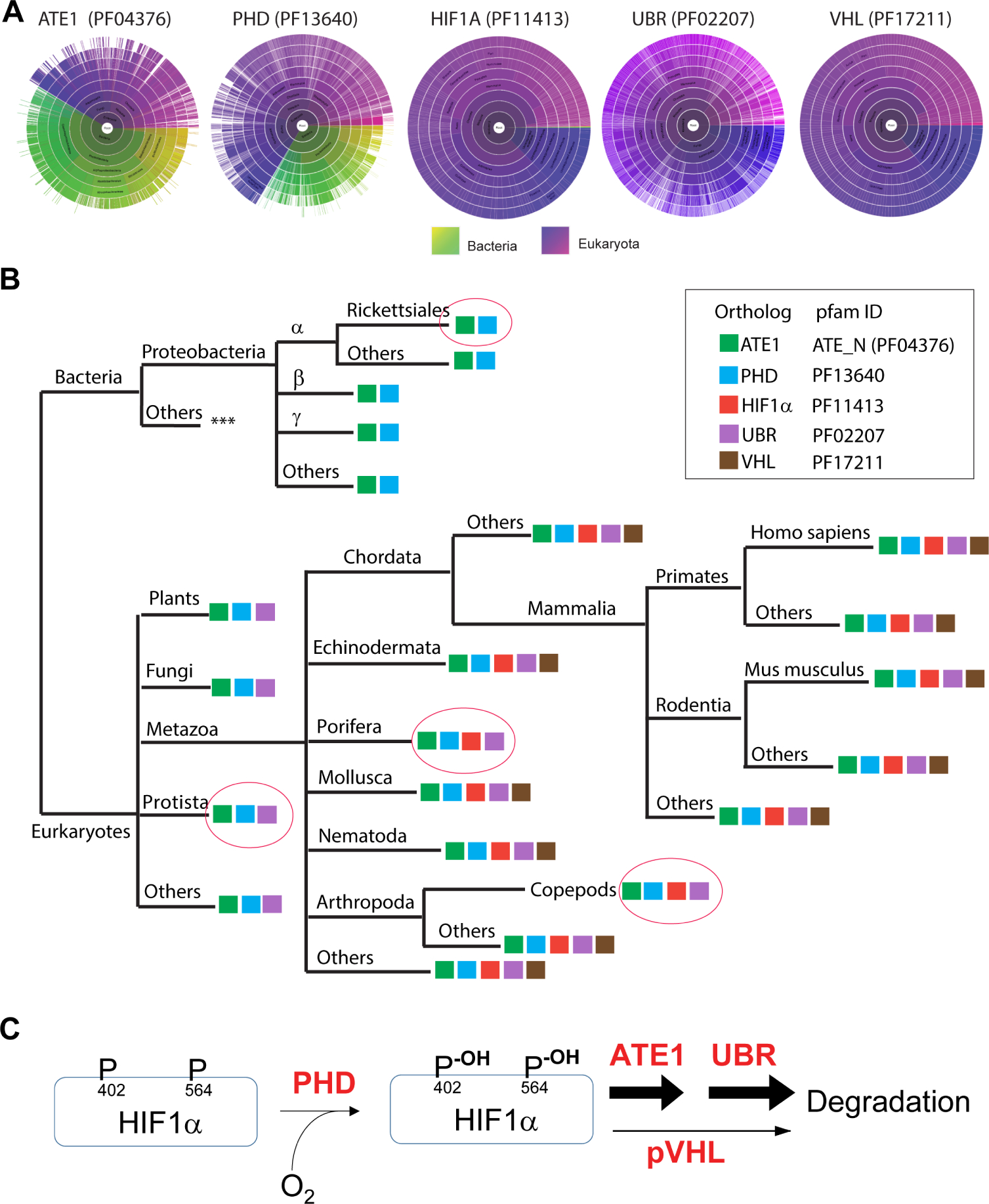
A) Sunburst graph showing the distribution of essential domains of ATE1, PHD, HIF1α, Ubr, and pVHL among eukaryotes and/or bacteria (indicated by color keys). For ATE1, the ATE-N domain (Pfam ID: 04376), which was known to be essential for its arginylation function(Kumar et al., 2016), was used as the representative. For PHD, the 2OG-Fe(II) oxygenase superfamily domain (Pfam ID: 13640), essential for catalyzing oxidation, was used. For HIF1α, Pfam ID 11413 was used. For VHL, the VHL box domain (VHL-C, Pfam ID: 17211) – required for the binding of cullin 2 protein, and thus for the ubiquitination of HIF1α – was used. For UBR, the zf-UBR domain (Pfam ID: PF02207), which is conserved among Ubr family, was used.
B) The presence of key Pfam domains of ATE1, PHD, HIF1α, pVHL, and Ubr (as indicated in the box) among different species are shown in a simplified evolution tree. Pink circles highlight several important spots. Among these, the Rickettsiales order of the alphaproteobacterial class – considered to be an extant relative to the ancestor of mitochondria – contains orthologs of ATE1 and PHD (e.g., GenBank: MBO88368.1 and GenBank: MBB20576.1, respectively, in https://www.ncbi.nlm.nih.gov/protein). Protista – considered a close relative to the ancestor of eukaryotes – contains ATE1, PHD and UBR. Further evidence can also be seen in Naegleria gruberi (Amoeba/Protista), where ATE1 (UniprotKB D2V1S2), PHD (UniprotKB D2V646), and Ubr (UniProtKB - D2V5N7) are found. Porifera is considered a close to the ancestor of metazoans, but it appears to lack pVHL. As an example, in Amphimedon queenslandica (sponge/Porifera), ATE1 (UniprotKB- A0A1X7UG82), PHD (UniprotKB- A0A1X7V6W1), HIF1α-like [NCBI accession no. XP_011403284.1], and Ubr (UniProtKB - A0A1X7UYL9) are found, but not for pVHL. A lateral loss of pVHL was also observed in Copepods (a branch of Arthropoda). As an example, in Eurytemora affinis (Copepods), we found ATE1 (NCBI accession no. XP_023347556.1), PHD (NCBI accession no. XP_023332905.1), HIF1A-like (NCBI accession no. XP_023346906.1), and Ubr (NCBI accession no: XP_023321273.1) but not pVHL. See Suppl Fig. S7A and S7B for sequences of the HIF1α-like proteins in Porifera and Copepods with putative arginylation-eligible residues. See also the figure legend of Suppl Fig. S7A and S7B for further details of the lack of VHL proteins in Protista, Porifera and Copepods.
C) The data in this study suggest that ATE1 acts downstream of PHD and parallel to pVHL for the degradation of HIF1α during oxygen sensing in mammalian cells.
