Figure 5.
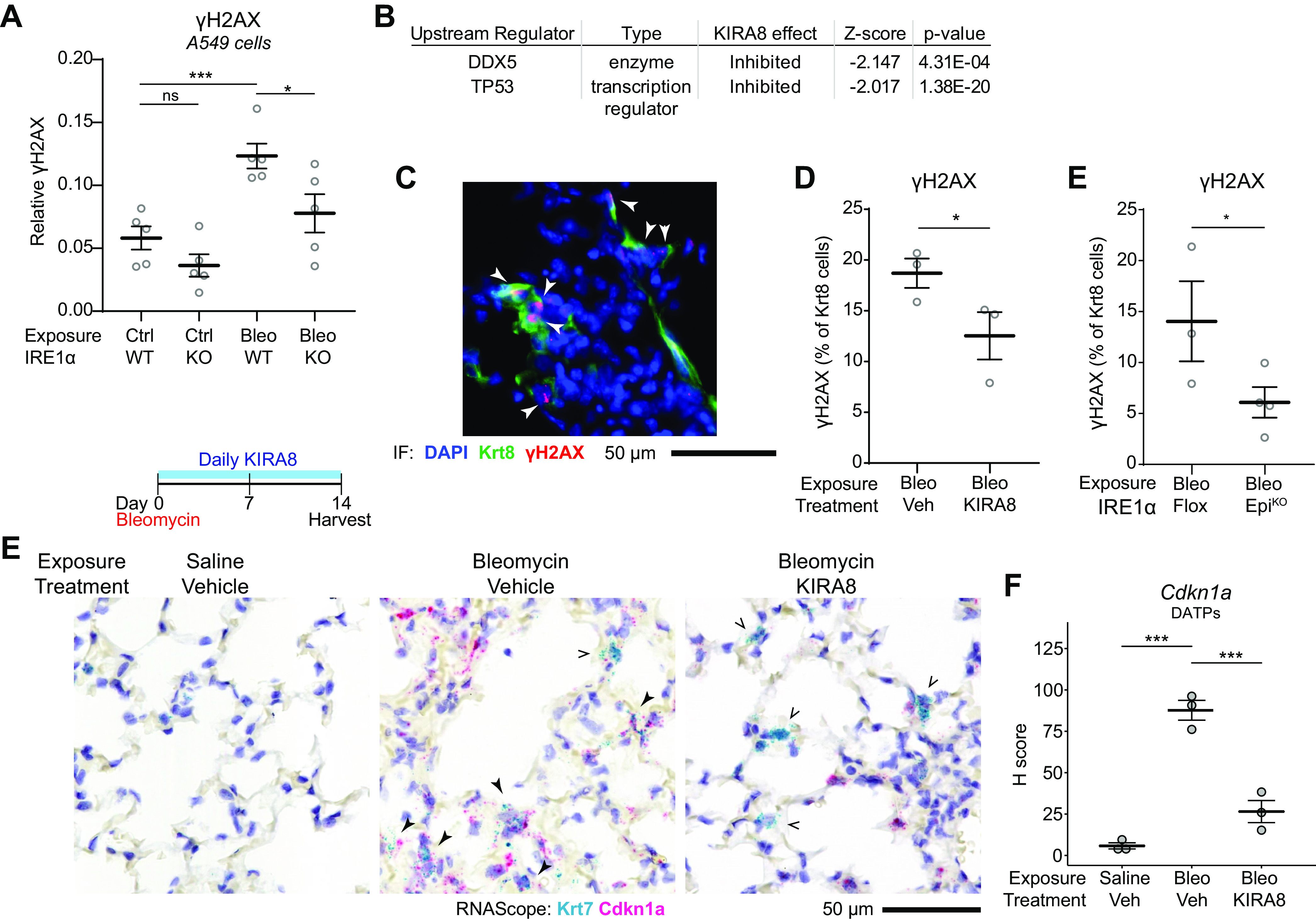
IRE1α reinforces DNA damage response signaling in DATPs. A: quantification of γH2AX expression in A549 cells or CRISPR-knockout IRE1α A549 cells. Cells were exposed to 5 µg/mL bleomycin for 4 h. Each dot represents an independent well with means ± SE indicated. Groups were compared by ANOVA with Fisher’s post hoc test. B: naïve upstream regulator analysis using the Ingenuity Pathway Analysis package on epithelial RiboTag data from bleomycin-exposed mice as in Fig. 1I, identifies p53 and DDX5 as inhibited DNA damage response pathways after KIRA8 treatment. C: representative immunofluorescence staining for Krt8 (green) and γH2AX (red) in mice exposed to bleomycin for 14 days. Arrowheads denote Krt8+ γH2AX double-positive cells. Automated counting of γH2AX and Krt8+ cells and quantification of the percentage of Krt8+ cells that are also γH2AX positive in mice treated with KIRA8 (D) or mice with conditional epithelial IRE1α knockout (E). Each dot represents the average of eight low power fields from one mouse with means ± SE indicated. Groups were compared by one-sided Student’s t test. E: RNA in situ hybridization (RNAscope) for Krt7 (teal) and Cdkn1a (red) in mice exposed to bleomycin and treated with daily KIRA8 for 14 days. Solid arrowheads denote Krt7 and Cdkn1a double-positive cells while arrow outlines indicate Krt7+ cells without significant Cdkn1a expression. F: semiquantitative H-score of Cdkn1a puncta in Krt7+ DATPs, averaged over 10 representative fields per dot with means ± SE indicated. Groups were compared by ANOVA with Fisher’s post hoc test. *P < 0.05, ***P < 0.001. DATP, damage-associated transient progenitor; KIRA8, kinase inhibitor of IRE1α.
