Figure 5. Selective reduction in Type 2 nRT mEPSC frequency in DS mice.
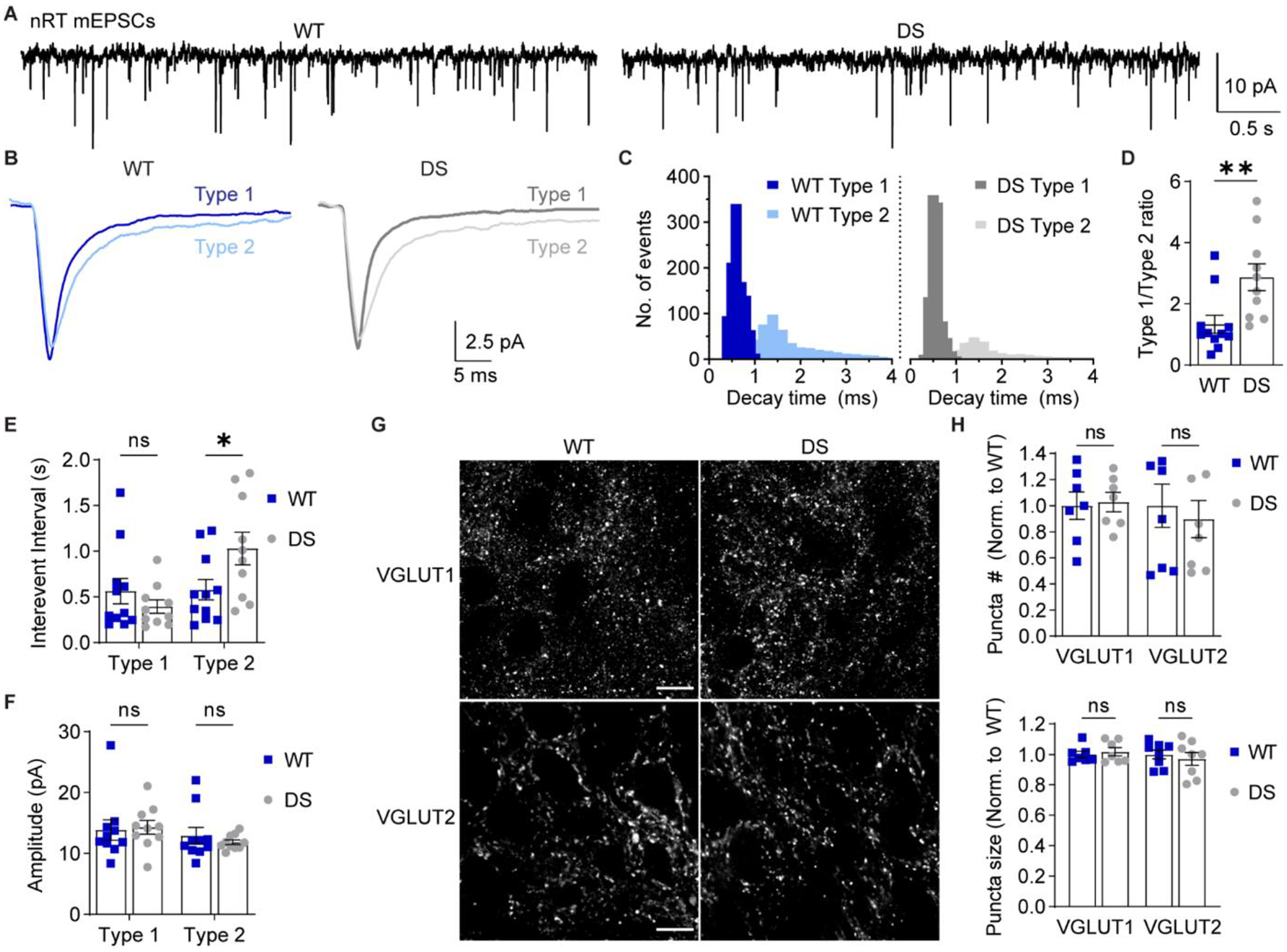
A. mEPSCs were recorded from nRT neurons in acute brain slices in the presence of TTX. B. Representative traces show ensemble averages of WT and DS Type 1 and Type 2 mEPSCs as determined by decay time. C. The decay time distributions from each cell were averaged for WT and DS groups to generate the depicted histogram of the mean number of events per cell (bin size = 0.1 ms). D. The ratio of Type 1/Type 2 events for WT (n = 11 cells from 6 mice) and DS (n = 10 cells from 6 mice) neurons were compared by unpaired t-test (*p = 0.008). E. Mean inter-event interval and (F) amplitude values for Type 1 and Type 2 mEPSCs were analyzed by two-way ANOVA with posthoc Sidak’s tests. Inter-event interval: Genotype F(1,38) = 1.169, p = 0.286; Interaction F(1,38) = 5.619, p = 0.023; Type 1 WT vs. DS: p = 0.600; Type 2 WT vs. DS: *p = 0.039. Amplitude: Genotype F(1,38) = 0.0699, p = 0.795; Interaction F(1,38) = 0.388, p = 0.538. See Supplementary Figure S3 for cumulative distributions of inter-event interval and amplitude data. G. Representative 100X images depict VGLUT1 and VGLUT2 staining in WT and DS nRT. Scale bar: 10 μm. H. The number and size of VGLUT1 and VGLUT2 puncta (n = 7 mice) were analyzed by two-way ANOVA. Puncta number: Genotype: F(1,26) = 0.081, p = 0.78; Interaction: F(1,26) = 0.250, p = 0.620. Puncta size: Genotype: F(1,26) = 0.031, p = 0.86; Interaction: F(1,26) = 0.545, p = 0.47. Data points in the bar graphs represent individual neurons (D-F) or mice (H).
