RNA is distinct among large biomolecules in that it has both informational coding ability, carried in its sequence, and the ability to form complex three-dimensional structures that can have catalytic and regulatory roles. The information-carrying component is widely appreciated. The pattern of base pairing—the first level of RNA structure—can be experimentally assessed and modeled with impressive accuracy (1, 2). By contrast, our understanding of the extent and roles of complex three-dimensional RNA structures remains rudimentary. RNA viral genomes are rich in motifs with complex three-dimensional structures with regulatory functions (3), and evidence increasingly supports the hypothesis that functional RNA structures are ubiquitous in organisms ranging from bacteria to humans. However, developing and testing hypotheses about the roles of RNA structure have been hindered by the inability to identify and model these structures. On page 1047 of this issue, Townshend et al. (4) report a machine-learning strategy for identifying native-like RNA folds.
Nearly all RNAs that form well-understood complex structures fall into a small number of classes: the ribosomal RNAs, the large and small ribozymes that catalyze RNA cleavage, bacterial riboswitches, and regulatory elements encoded by RNA viruses. Thus, there are limited examples for guiding identification and modeling of RNAs with complex three-dimensional structures. There are only four major RNA nucleotides, and the interactions that govern base pairing and simple helix formation are well understood. Once formed, RNA helices (secondary structure) often assemble as fairly rigid elements that interact hierarchically to form more complicated structures (tertiary structure) (see the figure). Despite these simplifying features, the modeling of complex RNA structures has proven to be difficult.
Figure. RNA structure.
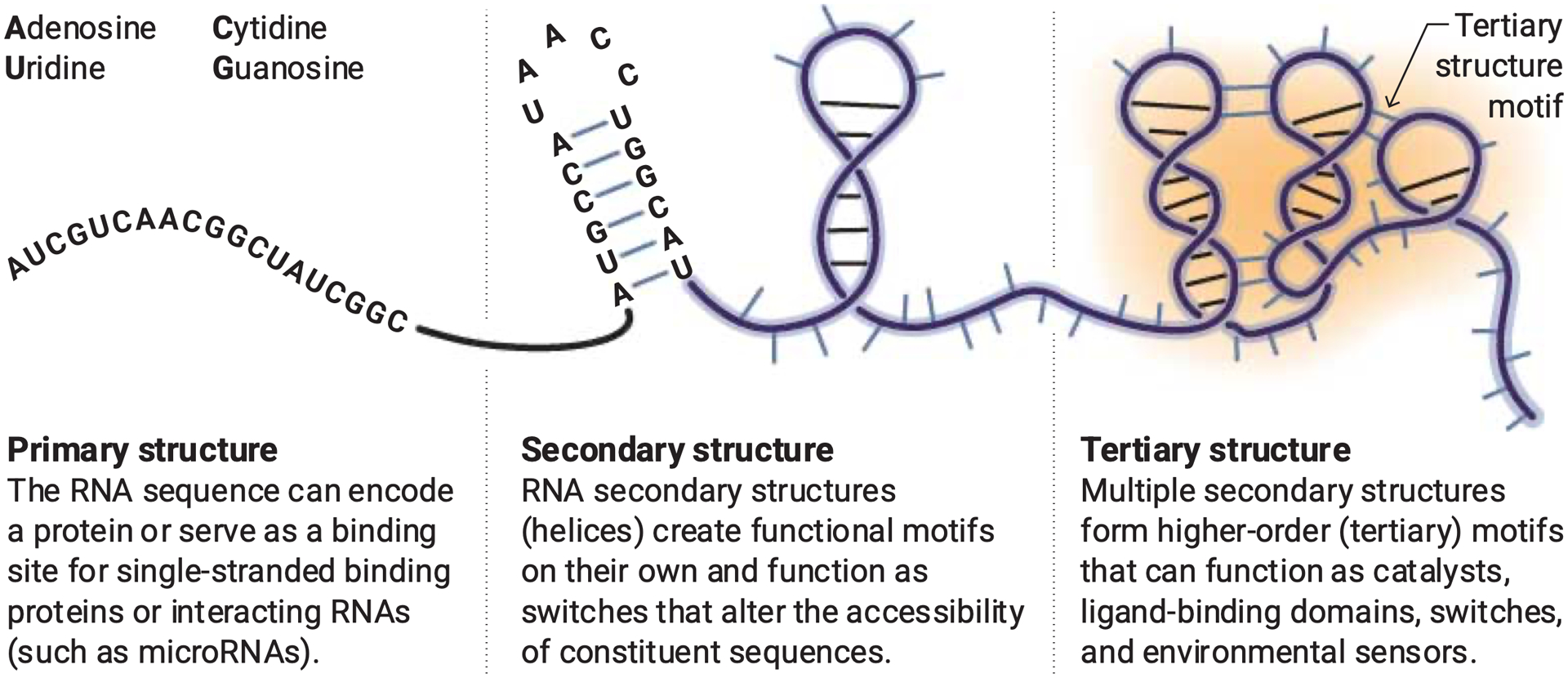
RNA molecules have multiple levels of structure and ability to encode information. The sequence of RNA is readily determined. RNA secondary structure can now be elucidated with high levels of accuracy using approaches that meld computational energy minimization with experimental per-nucleotide chemical probing information. Townshend et al. developed a deep neural network that can identify models that best represent the native tertiary state, taking a step toward modeling three-dimensional RNA structure.
The RNA-Puzzles community exercise (5, 6) has been instrumental in illuminating the challenges involved: Groups try to predict an RNA structure from its sequence before learning the solved structure. Several rounds of RNA-Puzzles have revealed important themes. No single method consistently yields the best models, although certain approaches have better records than others, and most approaches are getting better. The best agreement tends to result when experimental or homology-based information is incorporated into the computational modeling. However, the median accuracy for small RNAs, with complex tertiary folds but without a close known homolog, has stayed stubbornly stuck in a range of ~15- to 20-Å root mean square deviation [(RMSD) a measure of the similarity between known and modeled structures]. This agreement is much poorer than that now achieved for protein structures by machine learning (7), where native-like folds (~2-Å RMSD or less) are achieved. Modeled RNA structures thus often recapitulate the overall fold of a target RNA but do not consistently reveal details of the tertiary structure. Current methods are not likely to be useful for applications such as understanding the biological mechanism of a structure or for designing ligands (or drugs) that modulate RNA function.
The Atomic Rotationally Equivalent Scorer (ARES) approach of Townshend et al. is a deep neural network, a form of machine learning, and did not initially include preconceived notions of RNA structure. Indeed, the ARES framework is not specific to RNA and can be applied to other problems in molecular structure. Instead, ARES was given a small set of motifs with known RNA structure plus a large number of alternative (incorrect) variations of these same structures. ARES parameters were adjusted so that the program learned the functional and geometric arrangements of each atom and how these elements are positioned relative to each other. Layers in the neural network compute features from finer to coarser scales to recognize base pairs, helices, and more-complex structures. For example, ARES learned patterns of base pairing, the optimal geometry for RNA helices, and a subset of noncanonical tertiary motifs without being provided explicit information about these features of RNA structure.
Although ARES was trained on very simple RNA systems, the resulting ARES scoring function was able to predict structures of more complex RNAs, on average, to roughly a 12-Å RMSD. This degree of accuracy represents an overall improvement of ~4 Å over prior scoring methods. ARES is still short of the level consistent with atomic resolution or sufficient to guide identification of key functional sites or drug discovery efforts, but Townshend et al. have achieved notable progress in a field that has proven recalcitrant to transformative advances.
There are three fundamental challenges for modeling complex RNA three-dimensional structures: generating reasonable structures that may represent a biological state, accurately scoring or identifying models that best represent the correct native state, and using these hopefully accurate models to discover new functional motifs and to develop hypotheses regarding the mechanisms by which RNAs with complex three-dimensional structures regulate biological processes. The ARES machine-learning approach addressed the second of these three challenges: Candidate structures still need to be generated for evaluation by ARES. With further development, deep learning strategies hold promise for creating new scoring functions that can guide structure generation in ways that might yield near-native structures. Another important goal is to use a machine-learning strategy to identify regions in large RNAs most likely to fold into three-dimensional structures.
Current computational-only algorithms are not able to predict the pattern of base pairing in large RNAs accurately, even though base pairs are simpler to predict than tertiary structure. However, secondary structures for large RNAs are routinely modeled to high accuracies by incorporating experimental information. New, efficiently executed experiments are now being developed that measure features of RNA tertiary structures. Another frontier, analogous to recent advances in secondary structure modeling, would thus be to incorporate experimental information into machine-learning strategies for modeling RNA tertiary structure.
Large-scale investigation of RNA structure to date, primarily focused on RNA secondary structure, has revealed several core principles. One is that the existence of regions within large RNAs with complex, higher-order structure is unremarkable. When these base pairing and tertiary structures affect biological functions, they create “an RNA structure code” with pervasive effects on gene regulatory circuits. Additionally, every RNA likely has a distinct structural personality, which implies that there are numerous ways by which RNA structure tunes the underlying function of an RNA. At the level of secondary structure, such tuning RNA structures tend to function like switches and attenuators that modulate binding by RNA and protein ligands (8–11). Finally, characterization of well-determined RNA secondary structures often leads to identification of centers of new biology. As it becomes possible to measure, (deeply) learn, and predict the details of the tertiary RNA structure-ome, diverse new discoveries in biological mechanisms await.
Acknowledgments
The author’s laboratory is supported by the US National Institutes of Health and National Science Foundation. The author is an advisor to and holds equity in Ribometrix.
REFERENCES AND NOTES
- 1.Strobel EJ et al. , Nat. Rev. Genet 19, 615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weeks KM, Acc. Chem. Res 54, 2502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaafar ZA, Kieft JS, Nat. Rev. Microbiol 17, 110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Townshend RJL et al. , Science 373, XXX (2021). [Google Scholar]
- 5.Cruz JA et al. , RNA 18, 610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao Z et al. , RNA 26, 982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pennisi E, Science 373, 262 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Long D et al. , Nat. Struct. Mol. Biol 14, 287 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Kertesz M et al. , Nat. Genet 39, 1278 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Dominguez D et al. , Mol. Cell 70, 854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mustoe AM et al. , Biochemistry 57, 3537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


