Figure 2. Loss of function variants in PUS7 increases total protein translation.
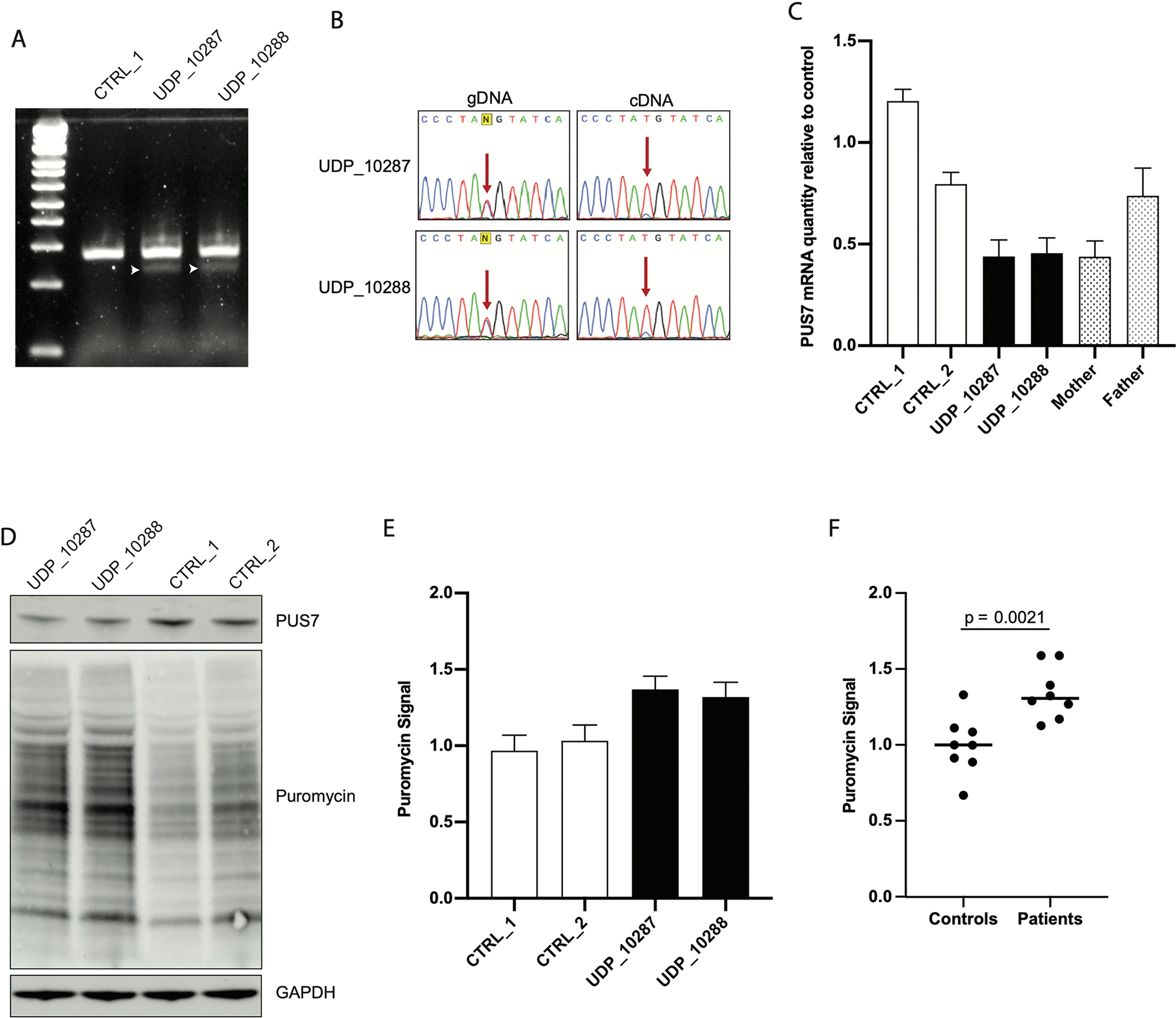
A) RT-PCR of RNA extracted from skin fibroblasts demonstrates the presence of a smaller splice form of PUS7 not detected in control. B) Sanger sequencing of RT-PCR products across the site of T387M shows that the majority of stable PUS7 mRNA is derived from the paternal allele. C) qRT-PCR of total RNA extracted from skin fibroblasts of the two patients, two age matched controls, and both parents demonstrates decreased PUS7 mRNA in individuals carrying the c.398+1 G>T splice variant (N = 9 for each sample); bar plots represent mean ± SEM. D) Western blots of total protein lysates purified from patient fibroblasts demonstrate that patients have decreased PUS7 protein quantity than controls and increased rate of protein translation relative to controls as measured by puromycin incorporation. E) Quantification of SUnSET assays conducted in fibroblasts (N = 4 for each sample); bar plots represent mean ± SEM. F) Combined analysis of SUnSET assay comparing control to patient fibroblast demonstrates statistically significant elevation of puromycin signal in patient cells (p = 2.1×10−3); bars represent mean value.
