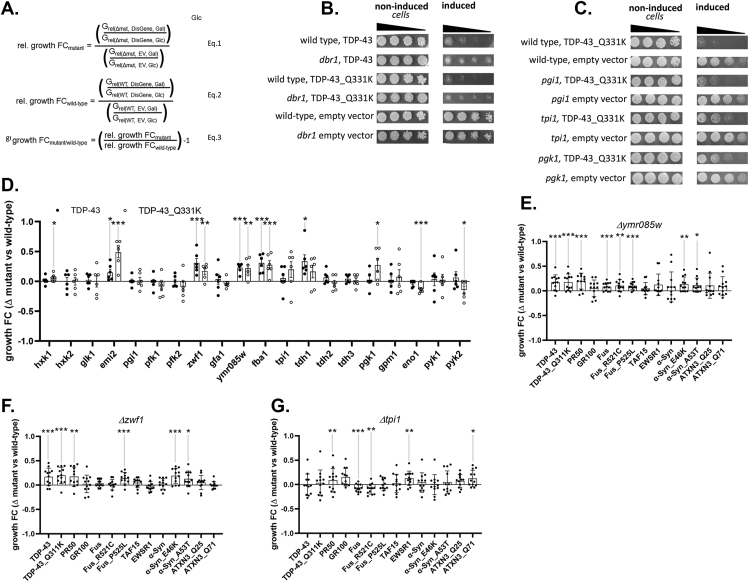
Figure 4.
Systematic deletion of individual glycolytic enzymes reveal modifiers of protein toxicity of neurodegenerative disease-associated proteins in yeast. Panel A. Equations used in semi-quantitative growth fitness assay consider three variables: 1) the presence or absence of a specific metabolism gene (noted as Wt or Δmut), 2) the presence of the empty Expression Vector or Disease Gene elaborated by the expression vector (noted as EV or DisGene), and 3) growth in Galactose or Glucose (noted as Gal or Glc). Note, the DisGene is expressed under the control of a galactose inducible promoter, and DisGene expression is inhibited in the presence of glucose. Equation one (Eq. (1)) describes the relative growth fold changes (“rel. growth FC”) in the yeast strain mutant (e. g., lacking) a metabolism gene, expressing the disease gene or not and growing on galactose or glucose. Equation two (Eq. (2)) describes the rel. growth FC of yeast strain WT for the metabolism gene, expressing the disease gene or not and growing on galactose or glucose. Equation (3) describes the specific effect of the metabolism gene on DisGene growth fitness by dividing Eq. (1) by Eq. (2), yielding the growth FCmutant/wild-type. Panel B. As previously published [44], deleting dbr1 partly rescues Wt and Mt (Q331K) TDP43-induced protein toxicity in the corresponding yeast dilution models. Panel C. Representative examples of glycolytic enzyme deletion strains tested for effects on TDP43 toxicity. The strain deleted for pgi1 did not affect protein toxicity, whereas strains deleted for tpi1 or pgk1 reduced Mt TDP-43 toxicity. Panel D. Screening deletion strains for glycolytic enzymes and for branching enzymes into PPP and HBS pathways using the Wt (filled circles) and Mt (open circles) TDP-43 protein toxicity models; n = 6. Growth fitness benefits (group FC > 0) were seen in yeast deleted for hxk1, emi2, zwf1, ymr085w, fba1 tdh1 and pgk1. Growth fitness decrement (growth FC < 0) were seen in yeast deleted for eno1 and pyk2. Panel E. Growth fitness benefits were seen in yeast deleted for ymr085w on a variety of proteotoxic insults relevant to ALS (e.g., TDP43, C9orf72 DPRs, FUS, TAF15, EWSR1) Parkinson's Disease (e.g., α-Syn) and Spinocerebellar Ataxia (e.g., ATXN3); n = 12. Growth fitness benefits were seen in yeast expressing Wt and Mt TDP43, PR50, FUS (Wt and P525L) and Wt and Mt (E46K and A53T) α-syn. Panel F. Growth fitness effects of ablation of zwf1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing Wt and Mt TDP43, PR50, Mt FUS and Mt (E46K and A53T) α-syn. Panel G. Growth fitness effects of ablation of tpi1 on a variety of proteotoxic insults relevant to ALS, PD and SCA; n = 12. Growth fitness benefits were seen in yeast expressing PR50, Wt and Mt FUS, EWSR1 and ATXN3 Q71 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005).