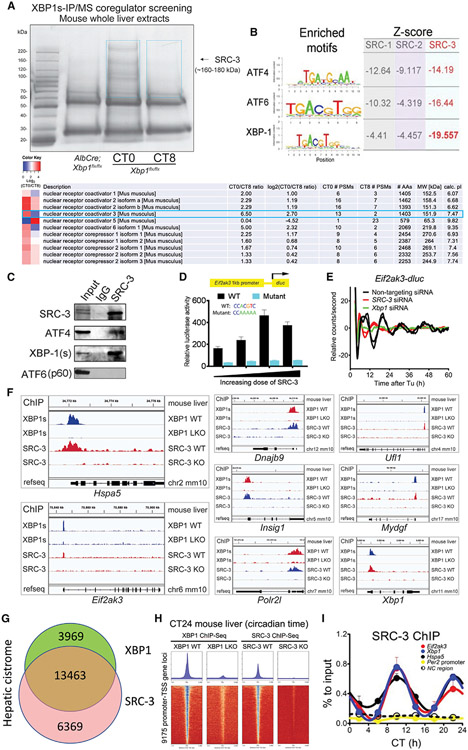
Figure 1. SRC-3 coactivates XBP1 transcription and regulates hepatic 12-h gene oscillation.
(A) Temporal XBP1s immunoprecipitation followed by mass spectrometry (IP/MS) coregulator screening in the mouse whole liver extracts identified SRC-3 as XBP1s-interacting protein candidate. Coomassie staining image of anti-XBP1s-immunoprecipitated proteins from the mouse liver extracts of Xbp1 flx/flx (WT) at circadian times CT0 and CT8, and AlbCre;Xbp1 flx/flx (LKO) mouse liver extracts are shown. SRC-3 identified by anti-XBP1s IP/MS is indicated with the black arrow. The temporal XBP1s-associated coregulators with CT0/CT8 ratio heatmaps peptide numbers, and length and molecular weight of the coregulators are shown. The two blue boxes show the gel slices submitted for MS analysis.
(B) Motif analysis of the hepatic cistrome of three major steroid receptor coactivators (SRCs) showed the strongest enrichment of XBP1 motif at SRC-3 binding sites. The enriched motifs of all three major transcriptional factors (TFs) ATF4, ATF6, and XBP1 for unfolded protein response (UPR) and the calculated Z-scores of all three major SRCs SRC-1, SRC-2, and SRC-3 are shown.
(C) Western blot (WB) verification of the specific binding of endogenous XBP1s (spliced form of XBP1) and ATF4, but not ATF6 (p60), to SRC-3 by anti-SRC-3 IP in the mouse liver. IgG was used as an IP control. The mouse whole-liver nuclei extracts prepared at CT10 were used in the IP and western blot assays. Western blotting was conducted with the antibodies specific to the indicated endogenous proteins.
(D) Transient luciferase assay using WT (XBP1s binding motif CACGTC) and XBP1s binding site mutant (a mutant promoter where CACGTC is converted to CAAAAA) Eif2ak3-dluc vector with increasing doses of SRC-3 expression vector in mouse embryonic fibroblast (MEFs) (n = 4).
(E) Real-time luminescence recording of non-targeting, SRC-3 siRNA- or Xbp1 siRNA-transfected Eif2ak3-dluc MEFs in response to tunicamycin (Tu) treatment. Detrended graphs are provided. For Tu treatment, two independent experiments are shown. Eif2ak3-dluc MEFs were transfected with different siRNAs and then shocked with a Tu treatment and real-time luciferase were recorded by Lumicycle for a total of 60 h.
(F–H) Representative binding profiles (F), Venn diagram (G), and heatmaps (H) of XBP1s and SRC-3 ChIP-seqs at the transcription start sites (TSSs) of 12-h-oscillating genes in the indicated mouse livers at CT24.
(I) ChIP-qPCR of hepatic SRC-3 on selective 12-h cycling gene promoters (Eif2ak3, Xbp1, and Hspa5), 24-h gene Per2 promoter, and noncoding (NC) region every 4 CT hours for 24 h in total (n = 3 mouse livers per time point).