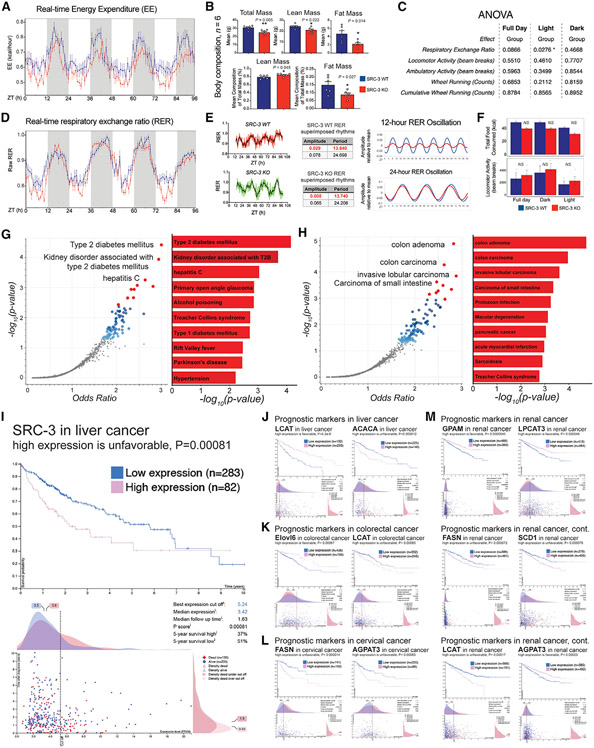
Figure 6. SRC-3 coactivation of 12-h clock regulates energy expenditure and links to metabolic diseases and cancer.
(A) The relative real-time energy expenditure (EE) values of the indicated mice (n = 4) housed under ad libitum feeding conditions. Averaged real-time EE values of the indicated mouse strains plotted against the indicated time points.
(B) The body composition of the indicated mice (n = 6) housed under ad libitum feeding conditions. Unpaired Student’s t test was performed with p value indicated.
(C) The integrated statistical analysis is shown for the indirect calorimetry experiments of the SRC-3 WT and KO mice (n = 4) housed under ad libitum feeding conditions. One-way ANOVA analysis with Tukey’s post hoc analysis was performed with p value threshold 0.05. The ANOVA was normalized by lean mass at the indicated light, dark, and full-day conditions (n = 4).
(D) The relative real-time respiratory exchange ratio (RER) values of the indicated mice (n = 4) housed under ad libitum feeding conditions. Averaged real-time RER values of the indicated mouse strains plotted against the indicated time points.
(E) Raw, eigenvalue/pencil decompositions, and deconvolutions of the mean RER oscillations of 12-h (top, in red) and 24-h (bottom, in black) oscillations in the SRC-3 WT and KO mice.
(F) Food intake (top) and locomotor activity (bottom) in the SRC-3 WT and KO mice fed regular chow ad libitum (n = 4). Blue line,: SRC-3 WT; red line, SRC-3 KO. Mouse RER, EE, body mass, food intake, and locomotor activity data are graphed as means ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001.
(G and H) Top enriched disease perturbations identified from the 486 concordant 12-h-oscillating genes regulated by both XBP1 and SRC-3. disease perturbations identified from downregulation (G) and upregulation (H) of the concomitant 486 12-h-oscillating genes are shown. Blue and red dots represent all the significant disease perturbations at discovery p value < 0.05. Top 10 disease perturbations are labeled as red dots.
(I–L) Survival outcome analysis of SRC-3- and SRC-3-regulated 12-h lipid metabolic genes ACACA, FASN, ELOVL6, SCD1, LPCAT3, LCAT, GPAM, and AGPAT3 in the indicated cancer types using TCGA pan-cancer clinical data resource. Clinical survival outcomes, survival scatterplot, cut-off threshold, median expression and follow-up time, p score, and 5-year survival percentage with high- and low-expression patient numbers are shown. Individual genes and their prognostic outcomes are indicated with specific cancer types.