Figure 16.
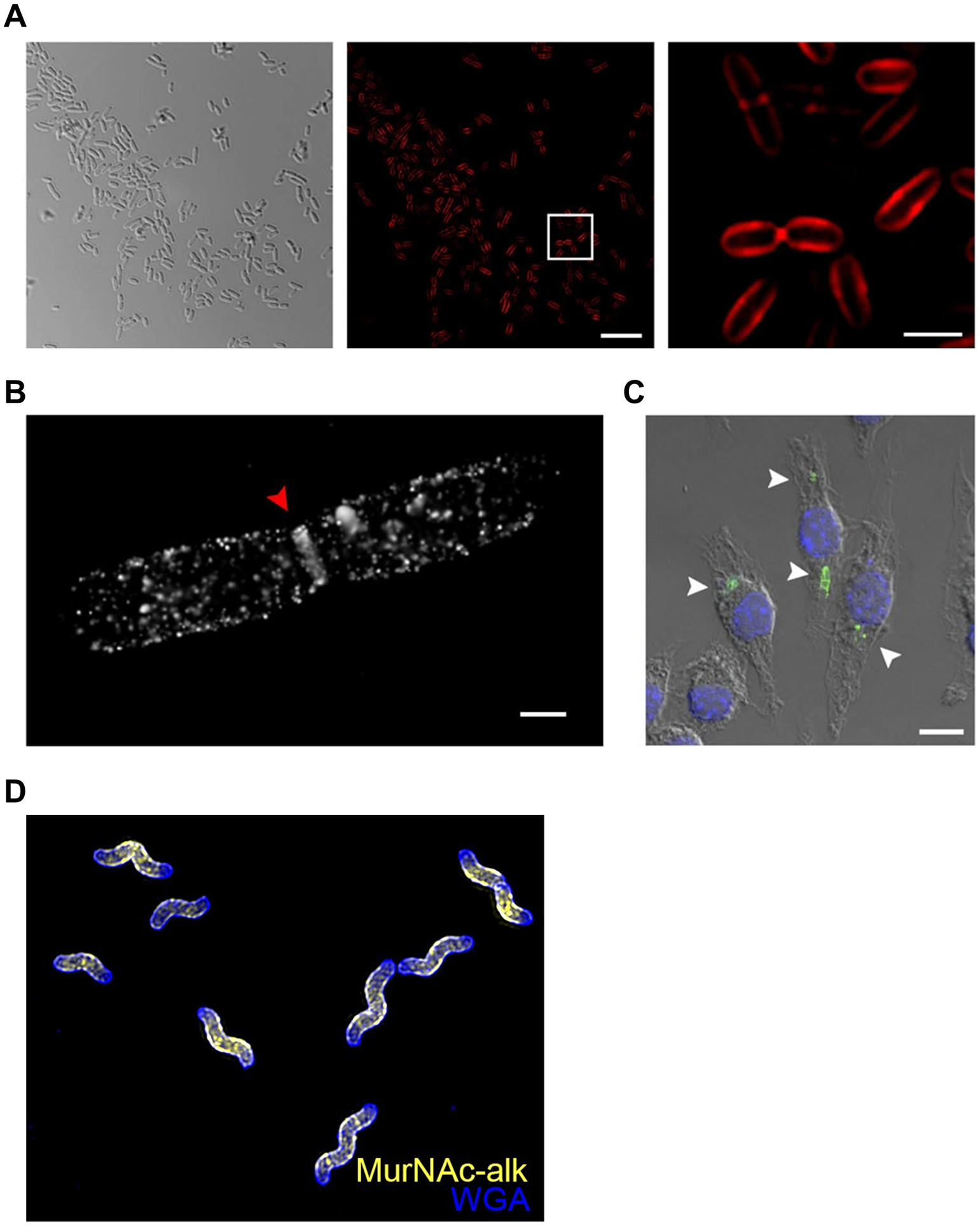
Imaging the PG glycan backbone using MurNAc reporters. (A) E. coli ΔMurQ-KU was incubated in the presence of alkynyl MurNAc 42 and fosfomycin, fixed, subjected to CuAAC with azido-Cy5, and imaged by structured illuminated microscopy (SIM). Left, differential interference contrast (DIC); middle, SIM image; right, boxed area from middle image. (B) E. coli ΔMurQ-KU treated as in (A) and imaged by super-resolution 3D stochastic optical reconstruction microscopy (STORM). Red triangle indicates septal division plane. (C) E. coli ΔMurQ-KU were pre-treated with 42 and fosfomycin, then used to invade J774 macrophages. After fixation, CuAAC with azido-488, and nuclear staining with DAPI, invaded macrophages were imaged. White triangles indicate fluorescent fragments released from intracellular bacteria. Images in (A–C) are reproduced from ref 89. (D) H. pylori HJH1 was incubated in the presence of alkynyl MurNAc 42 for a short pulse, fixed, subjected to CuAAC with azido-Alexa Fluor 555, counterstained with the fluorescent lectin WGA-Alexa Fluor 488, and imaged by 3D SIM. Image in (D) reproduced from ref 154.
