Figure 41.
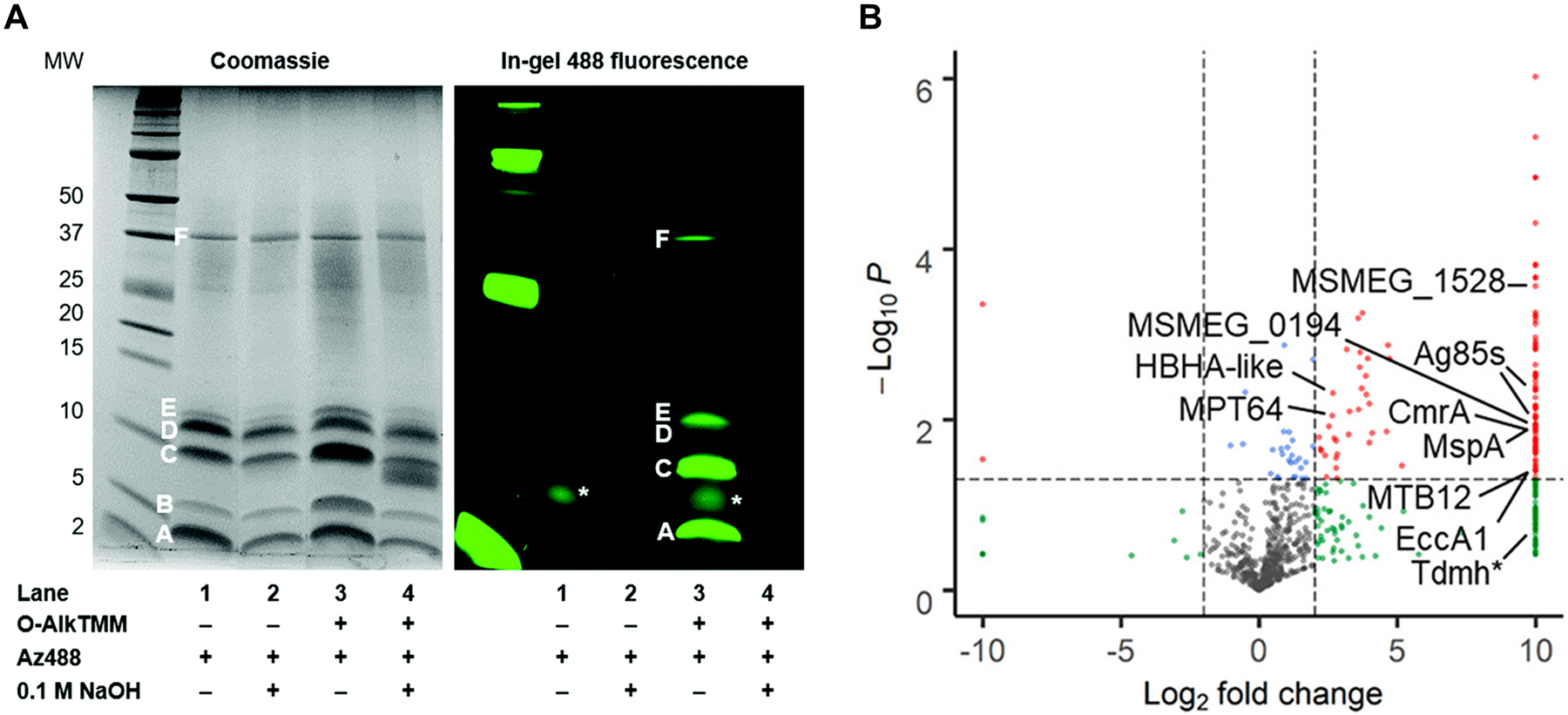
TMM derivatives enable labeling of proteins that (A) are covalently modified by or (B) non-covalently interact with mycolates. (A) Labeling of O-mycoloylated proteins in C. glutamicum with O-AlkTMM-C7. C. glutamicum was incubated in 100 μM O-AlkTMM-C7 (113), then chloroform-methanol protein extracts were obtained, subjected to CuAAC with azido-488, subjected to NaOH treatment to cleave ester linkages, and analyzed by SDS-PAGE. Bands A–F represent proteins identified by Coomassie staining; all but band B were labeled by O-AlkTMM-C7 and bands A, C, D, and E were confirmed by MALDI-MS to be O-mycoloylated. The asterisk (*) marks background fluorescence signal. Reproduced from ref. 332 with permission from the Royal Society of Chemistry. (B) Enrichment of mycomembrane proteins in M. smegmatis using photo-activatable N-x-AlkTMM-C15. M. smegmatis was incubated in 100 μM N-x-AlkTMM-C15 (122) and exposed to UV irradiation, then lysates were collected, subjected to CuAAC with azido-TAMRA-biotin, incubated with avidin beads, trypsinized, and analyzed by label-free quantitative LC-MS/MS. Volcano plot shows proteins in red that were significantly ≥4-fold enriched in 122-treated, UV-exposed versus non-UV-exposed bacteria. Selected proteins of interest are indicated. Reproduced with permission from ref. 332. Copyright 2020 American Chemical Society.
