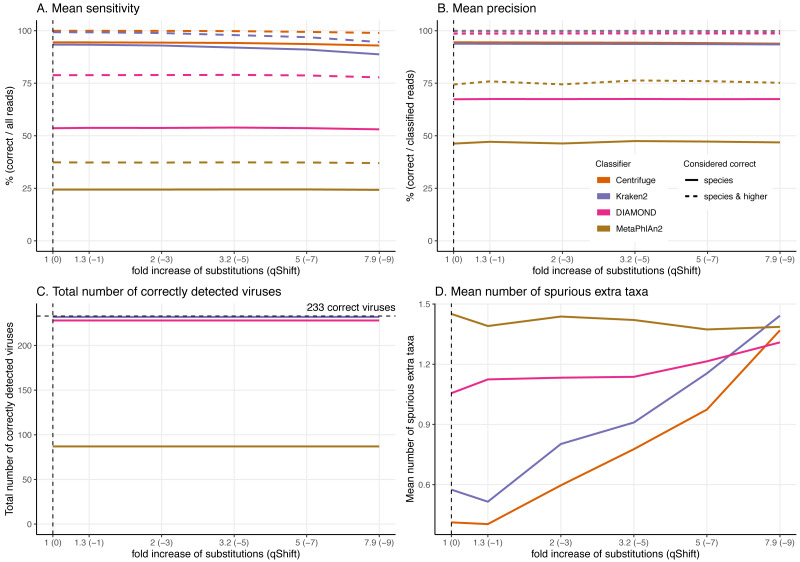
Figure 7. Effect of the substitution sequencing error on the classification performance.
For these simulations, errors were added using ART which assumes a profile similar to the ones observed for Illumina Sequencing machines (HiSeq 2500). The results shown here correspond to increasing the overall sequencing error rate ranging from 1 to 7.9-fold (qShift values from 0 to −9). On the x-axis, the first number correspond to the expected fold increase in error rate while the parameter that was varied, qShift, is shown in parenthesis. (A) Average Sensitivity_s (continuous lines) and Sensitivity_s&h (dashed lines) for each classifier. (B) Average Precision_s (continuous lines) and Precision_s&h (dashed lines) for each classifier. (C) Total number of viruses detected out of the 233 tested. The dashed line shows the maximum number of detectable viruses. (D) Average number of spurious extra taxa across simulated viral sequences. The vertical dashed line indicates the initial 60 bp read set.