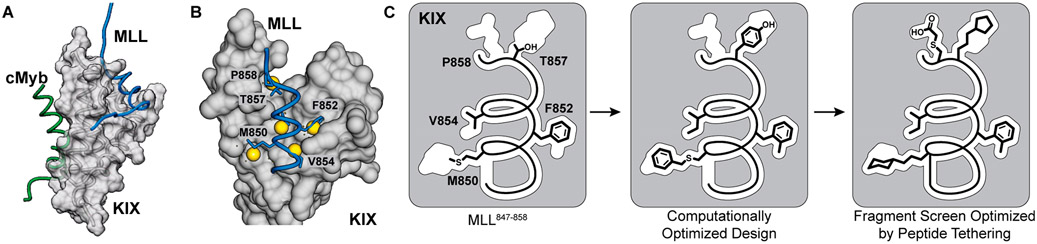
Figure 1.
(A) KIX domain of coactivators p300/CBP interacts with a multitude of transcription factors. An NMR-derived model of KIX in complex with MLL and cMyb is shown (PDB code 2AGH). (B) Helical domain of MLL847–858 provides a template for the development of synthetic ligands for KIX. The topographical map of KIX suggests that several pockets on its surface are not optimally occupied by native MLL residues, and nonnatural residues may be designed to provide enhanced affinity. The figure shows AlphaSpace analysis of the KIX/MLL complex. The yellow spheres depict the centroid of potential pockets near the MLL helix. (C) In published studies, we showed that a computationally designed peptide with noncanonical amino acids replacing M850, F852, V854, and T857 makes superior contacts with KIX. Here we build on the computational method and describe an experimental fragment screening approach to identify synthetic side chains to engage KIX.