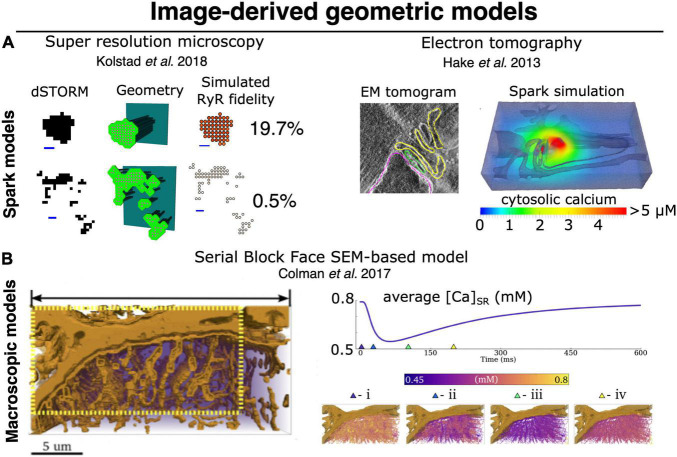
FIGURE 4.
Image-derived approaches to geometric modeling of CRU and whole-cell cardiac ECC. (A) Image-driven models of single dyad (and CRU) structures have been implemented beginning with both super resolution light microscopy (dSTORM), (left) (Walker et al., 2014; Kolstad et al., 2018), and tilt-section tomography from high voltage TEM imaging (right) (Hake et al., 2012; Edwards et al., 2018). To date, no correlative light/EM datasets visualizing both protein and membrane localization have been employed for this purpose, although such approaches would be highly desirable. (B) High-volume EM methods, particularly SBF-SEM, have recently been used as a basis for constructing image-driven geometries for much larger subsections of a cardiac myocyte (Colman et al., 2017; Colman, 2019). Simulations in these geometries can be used to interrogate the interaction of local Ca2+ release with uncertain (and difficult-to-measure) physiological properties, such as intra-SR Ca2+ diffusion. At right, spatially distributed intra-SR Ca2+ concentration is shown at various times during the evoked macroscopic SR Ca2+ release. Models of this type will likely provide the basis for bridging between the stochastic dynamics of single CRUs (and single RyRs) and pseudo-continuous dynamics of macroscopic CICR, thereby allowing us to better deconvolve the multitude of factors known to influence both. Copyright permission was not required to reproduce the figures.