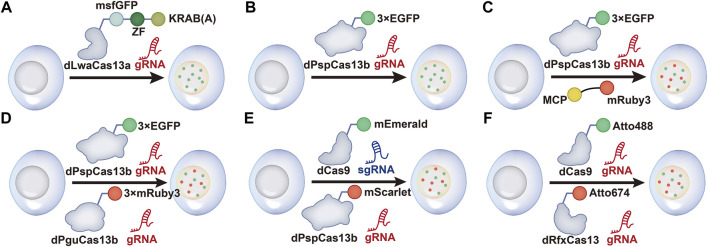
FIGURE 1.
Schematic of CRISPR/Cas13-mediated RNA imaging methods. (A) dCas13a-GFP-KRAB construction for negative-feedback imaging. The dLwaCas13a incorporates a negative-feedback system based upon zinc finger self-targeting and the KRAB repression domain to image ACTB mRNA. (B) dPspCas13b-3 × EGFP labeling system. dPspCas13b is tagged with several green fluorescent proteins EGFP to image mRNAs. (C) Dual-color RNA labeling using a combination of the dPspCas13b and MS2- MCP systems. A total of 24 × MS2 (24 copies of the MS2 stem loop)-NEAT1-KI HeLa cells are constructed. Then, RNAs are labeled simultaneously by transfecting dPspCas13b-3 × EGFP, gRNAs for NEAT1, and MCP-mRuby3 into the cells. (D) Dual-color RNA labeling by different dCas13b systems. RNAs are labeled with dPspCas13b and dPguCas13b in HeLa cells. (E) Dual-color labeling using dCas9 and dPspCas13b. DNA and RNAs are labeled simultaneously in living cells by combining the dCas9-mEmerald and dPspCas13b-mScarlet systems. (F) Dual-Color labeling using dCas9 and dRfxCas13. Atto488-labeled dCas9 and Atto647-labeled dRfxCas13 are used to image genomic DNA and RNA transcripts.