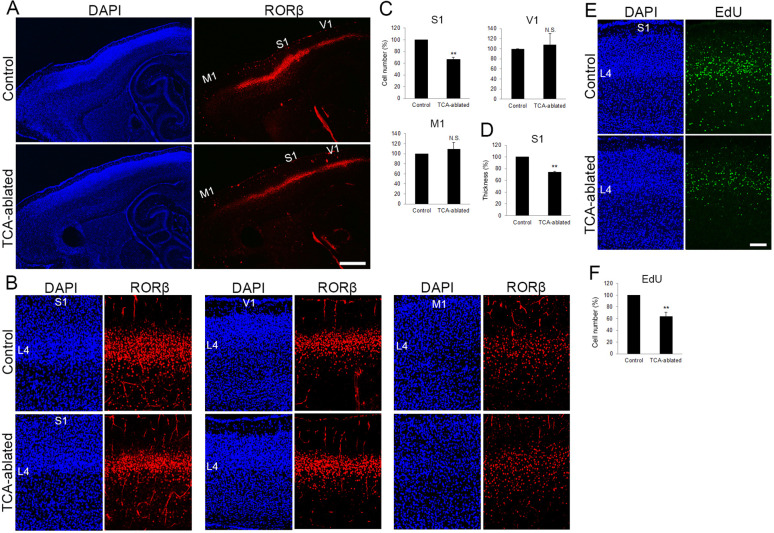
Figure 2. The number of layer 4 neurons is reduced in S1.
(A) Sagittal sections of P7 cortices derived from thalamocortical axon (TCA)-ablated mice stained with RORC antibody. Expression of the layer 4 marker RORβ was declined, specifically in S1, resulting in poorly demarcated borders between adjacent areas. (B) Coronal section of the cortex of a TCA-ablated mouse at P7, showing RORβ-expressing layer 4 (L4) neurons in S1. (C) Quantification of RORβ expression. RORβ-expressing cells within an 850-μm-wide strip of the cortical wall were counted. Note that the number of RORβ-positive cells was less in S1, but was not changed in V1 and M1, in TCA-ablated mice. Data are presented as a percentage of control mice (mean ± standard error of the mean [SEM]): S1, 66.75 ± 3.30%, N = 7 mice, p = 0.0011; V1, 108.28 ± 22.00%, N = 5 mice, p = 0.656; M1, 109.47 ± 12.97%, N = 4 mice, p = 0.878; Mann–Whitney U test, **p < 0.01. The same numbers of control and experimental animals were used. (D) Thickness of RORβ-expressing layer was also reduced in TCA-ablated mice. Data are presented as a percentage of control mice (mean ± SEM): 73.69 ± 1.83%, N = 7 mice, p = 0.0011; Mann–Whitney U test, **p < 0.01. (E) Cross-sections of S1 cortices of control and TCA-ablated mice at P7, showing distribution of EdU-positive cells. EdU was injected at E14.7. (F) Quantification of the results. The number of EdU-positive cells within an 850-μm-wide strip of the cortical wall relative to control is shown as the mean ± SEM: 64.37 ± 6.77%, p = 0.0075; Mann–Whitney U test, **p < 0.01, N = 5 animals for both control and TCA-ablated mice. L4, layer 4; M1, motor area; S1, primary somatosensory area; V1, primary visual area. Scale bars, 500 μm (A), 100 μm (B, E).
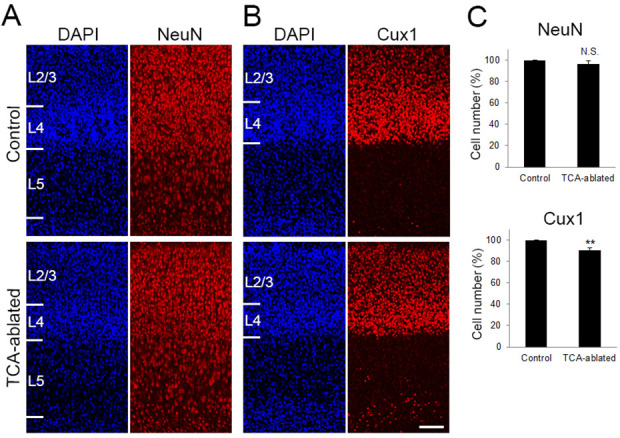
Figure 2—figure supplement 1. Effects of thalamocortical axon (TCA) ablation on the number of neurons in L2–5 and Cux1-positive upper layer neurons.