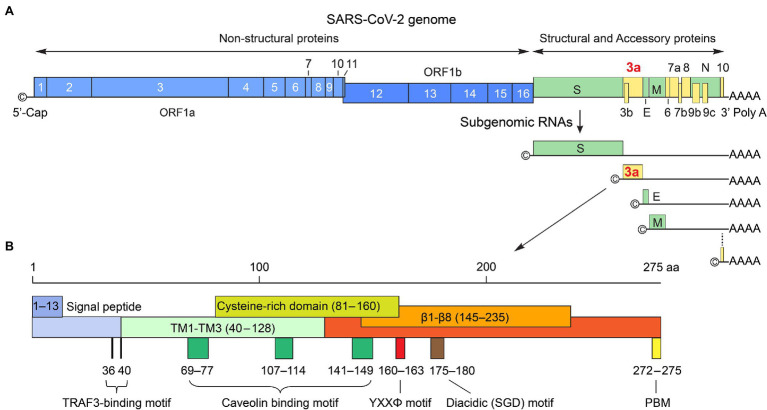
Figure 1.
SARS-CoV-2 genome organization and ORF3a protein. (A) SARS-CoV-2 RNA genome organization. The genome size is about 29.7 kb of non-segmented, +ssRNA. The 5′ cap structure and 3′ poly (A) tail are for translation of ORF1a and ORF1ab that are further processed to generate 16 non-structural proteins (NSP1-NSP16) that include all major replicase genes and enzymes that are needed for subsequent viral transcription and replication. The sgRNAs produced in RTC on DMVs are for 4 structural proteins (S, E, M, and N) and 9 accessary ORFs (3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10). These proteins are produced from a nested set of sgRNAs that share common 3′ ends and a common leader from the 5′ end. ORF3a is the largest accessory protein. It localizes between S and E as shown. (B) The nucleotide sequence of ORF3a is 825 bp in length and encodes a 31 kD protein of 275 a.a. It is a membrane-associated protein with an extracellular N-terminal (a.a. 1–39), 3 TM domains (TM1-TM3; a.a. 40–128); a short cytoplasmic loop (a.a. 175–180) with 8 β-sheets (β1–β8; a.a., 145–235) and a C-terminus (a.a. 239–275). Known and well-conserved functional motifs are depicted; details of these motifs are described in text, including an N-terminal signal peptide (a.a. 1–13); a TRAF3-binding motif (a.a. 36–40; Siu et al., 2019; Jin et al., 2021); a cysteine-rich domain (a.a. 81–160; Lu et al., 2006); 3 caveolin-binding motifs (a.a. 69–77, 107–114 and 141–149; Padhan et al., 2007); a YXXΦ motif (a.a. 160–163) and a diacidic (SGD) motif (a.a. 175–180; Tan et al., 2004; Minakshi and Padhan, 2014), and a PBM (a.a. 272–275; Castano-Rodriguez et al., 2018; Caillet-Saguy et al., 2021).