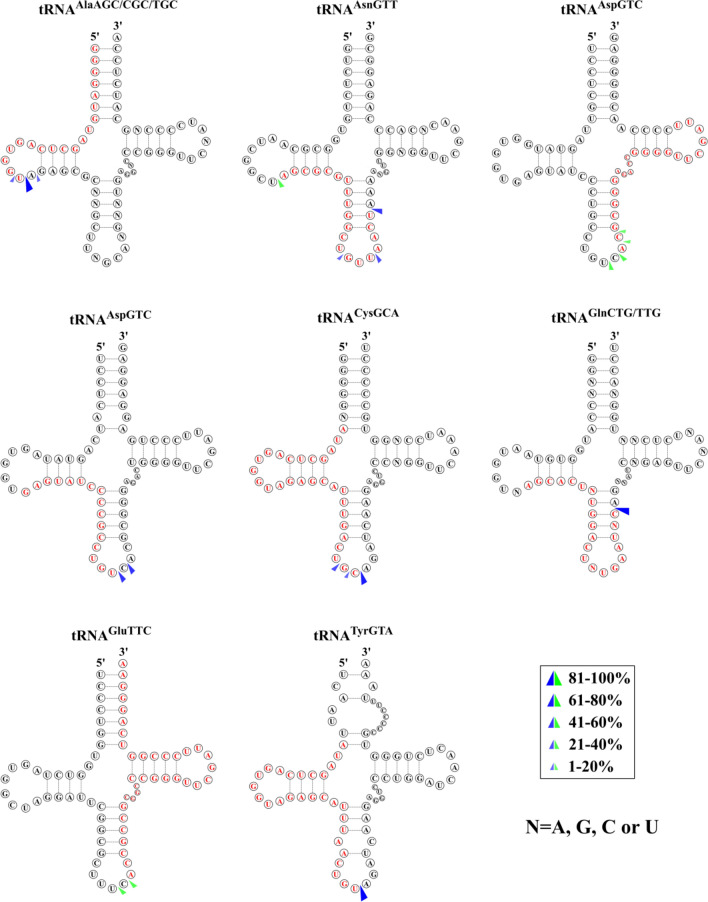
Fig. 4.
Predicted cleavage sites of the most significantly abundant tRNA-derived fragments identified by cP-RNAseq. Parental tRNAs with significant coverage differences between WT and RNase2-KO macrophage cells are depicted and the identified tRFs are marked in red. The possible cleavage sites are based on the 3′-terminal positions of the five prime fragments (blue arrows) and the 5′ terminal positions of three prime fragments (green arrows) according to the different coverage. Only sequences with a log2fold > 0.5 and adjusted p value < 0.05 are included