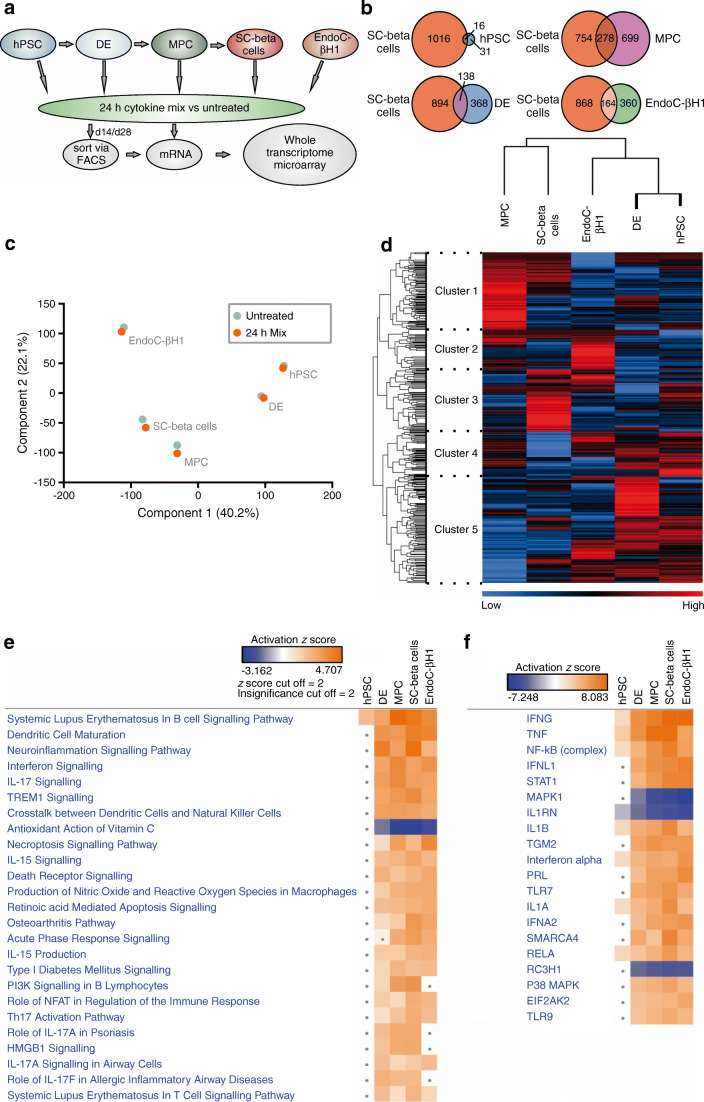
Fig. 3.
Analysis of transcriptome microarray data. (a) Schematic description of sample generation for the transcriptome analysis. (b) Number of genes jointly regulated (fold change ≥2) with SC-beta cells over the course of differentiation and in comparison with EndoC-βH1 cells, after a 24 h cytokine exposure. (c) PCA of the generated transcriptome data. (d) Hierarchical clustering of expression fold changes of cytokine-treated cells during the different stages of differentiation and EndoC-βH1 cells. (e) Comparative canonical pathway analysis of hPSCs (d0), DE (d4), MPCs (d14), SC-beta cells (d28) and EndoC-βH1 cells after cytokine treatment calculated by IPA. (f) IPA upstream analysis of cytokine-treated hPSCs, DE, MPCs, SC-beta cells and EndoC-βH1 cells. EIF2AK2, eukaryotic translation initiation factor 2 alpha kinase 2; HMGB1, high mobility group box 1; IFNA2, interferon alpha 2; IFNG, interferon gamma; IFNL1, interferon lamda 1; IL1A, interleukin 1 alpha; IL1B, interleukin 1 beta; NFAT, NFAT nuclear factor; PI3K,phosphatidylinositol 3-kinase; PRL, prolactin; RC3H1, ring finger and CCCH-type domains 1; RELA, RELA proto-oncogene, NF-kB subunit; SMARCA4, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4; TGM2, transglutaminase 2; Th17, T-helper 17; TREM1, triggering receptor expressed on myeloid cells 1