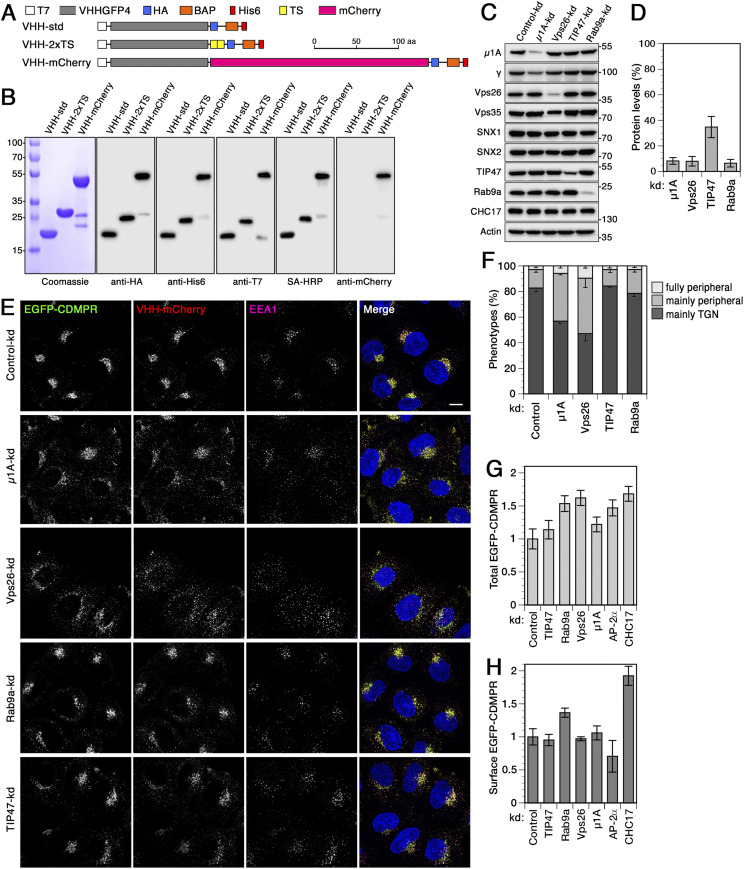
Figure 1. Functionalized nanobodies to analyze retrograde transport of CDMPR upon silencing of adaptor protein-1, retromer, Rab9a, or TIP47.
(A) Schematic representation of the functionalized nanobodies. The standard nanobody (VHH-std) consists of the GFP-specific VHH domain, T7 and HA epitope tags, a biotin acceptor peptide (BAP), and a hexahistidine (His6) purification tag. Other nanobodies in addition contain two tyrosine sulfation sequences (VHH-2xTS) or mCherry (VHH-mCherry). Scale bar in aa. (B) Bacterially expressed and purified nanobodies (30 μg) were analyzed by SDS-gel electrophoresis and Coomassie staining (left). Immunoblot analysis of nanobodies (10 ng) with antibodies against the HA, His6, T7, or mCherry epitopes, or with streptavidin-HRP (SA-HRP). Marker proteins with molecular weights in kilodalton are shown on the left. As previously reported (Buser et al, 2018; Buser & Spiess, 2019), mCherry-containing nanobodies are slightly susceptible to clipping between the VHH and mCherry domains. (C) HeLa cells were transfected with non-targeting siRNA or siRNAs targeting μ1A-adaptin, Vps26, TIP47, or Rab9a. 3 d after transfection, the cells were subjected to immunoblot analysis with antibodies against the indicated proteins. (D) To determine the knockdown (kd) efficiency, the residual protein was quantified in percent of the value after control-kd (mean and SD of three independent experiments). (E) HeLa cells stably expressing EGFP-CDMPR were depleted of μ1A-adaptin, Vps26, TIP47, or Rab9a as in (C). Cells were incubated for 1 h at 37°C with full medium containing 5 μg/ml VHH-mCherry (∼0.1 μM), fixed, stained for EEA1 and nuclei (DAPI, blue), and imaged by fluorescence microscopy. Bar: 10 μm. (F) Quantitation of the percentage of cells displaying the CDMPR localization phenotypes “mainly TGN,” “mainly peripheral,” or “fully peripheral” as in Wassmer et al (2007) and Simonetti et al (2017). For each condition, random frames with a total of 136–140 cells were scored from three independent experiments. (G, H) Total and surface EGFP-CDMPR levels in RNAi-silenced cells were quantified by flow cytometry. Cells were incubated for 30 min at 4°C with VHH-mCherry for exclusive binding to EGFP-CDMPR at the cell surface, washed, dissociated and analyzed for GFP and mCherry fluorescence to determine the levels of total (G) and surface EGFP-CDMPR (H), respectively. Median fluorescence intensities above background of parental HeLa cells without EGFP-CDMPR of each condition were normalized to the average of cells treated with non-targeting control siRNA. For each condition, 50,000 cells were analyzed in each experiment (mean and SD of three independent experiments).