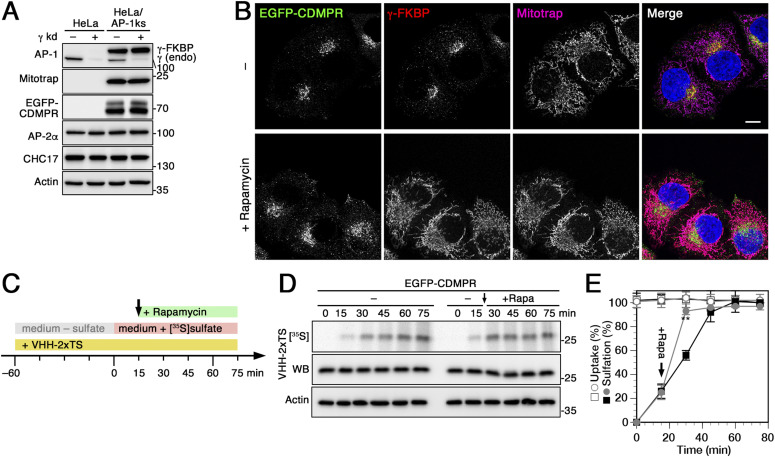
Figure 4. Increased nanobody sulfation upon adaptor protein (AP)-1 silencing is the consequence of anterograde TGN exit block.
(A) Lysates of normal HeLa cells and HeLa-AP1 knocksideways (HeLa-AP1ks) cells stably expressing γ-FKBP, Mitotrap and EGFP-CDMPR with or without siRNA-mediated knockdown of the endogenous γ-adaptin were subjected to immunoblot analysis for both forms of γ-adaptin, for Mitotrap (anti-FLAG), EGFP-CDMPR, the α-adaptin subunit of AP-2, and clathrin heavy-chain (CHC17). Knockdown efficiencies for endogenous γ-adaptin were typically >85%. (B) HeLa-AP1ks cells stably expressing EGFP-CDMPR after silencing endogenous γ-adaptin were treated with or without 500 nM rapamycin for 1 h and processed for fluorescence microscopy to detect EGFP-CDMPR, recombinant γ-FKBP (using an antibody targeting an epitope present in the neuronal splice variant of AP-2α), and Mitotrap (anti-FLAG). Bar: 10 μm. (C) Schematic outline of the anterograde transport sulfation assay in HeLa-AP1ks/EGFP-CDMPR cells. (D) HeLa-AP1ks cells stably expressing EGFP-CDMPR were siRNA-silenced for endogenous γ-adaptin, followed by starvation for sulfate in the presence of VHH-2xTS to preload all EGFP-CDMPR in the surface/endosome/TGN pool. The cells were then labeled with [35S]sulfate for up to 75 min in the continued presence of VHH-2xTS, without or with addition of 500 nM rapamycin after 15 min (arrow) to inactivate AP-1 (+Rapa). The nanobodies were isolated by Ni/NTA beads and subjected to SDS-gel electrophoresis followed by immunoblot analysis (anti-His6) and autoradiography ([35S]). In parallel, aliquots of the cell lysates were immunoblotted for actin as a control for the amount of cells used. (E) Three independent experiments as shown in panels C were quantified and presented as the percentage of the value in the absence of rapamycin after 75 min (mean and SD of three independent experiments; two-sided t test: *P < 0.05; **P < 0.01). Without rapamycin is shown as black squares, with rapamycin as gray circles; uptake as open symbols, sulfation as filled symbols.