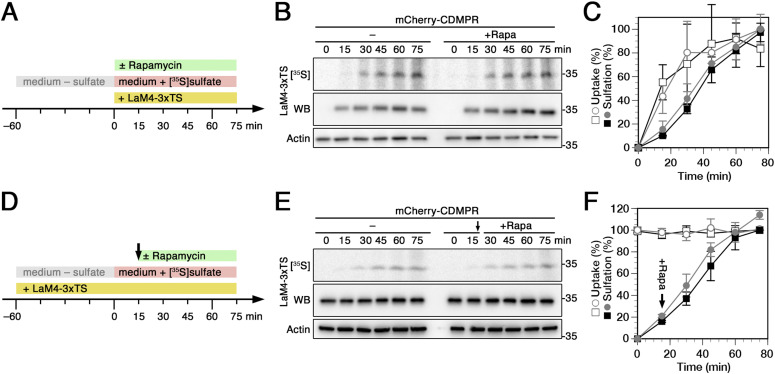
Figure 7. Effect of rapid GGA2 inactivation on retrograde and anterograde CDMPR transport using derivatized anti-mCherry nanobodies.
(A, D) Schematic outline of the retrograde (A) anterograde (D) transport sulfation assays, respectively, with HeLa-GGA2ks/mCherry-CDMPR cells. (B) HeLa-GGA2ks cells stably expressing mCherry-CDMPR were siRNA-silenced for endogenous GGA1–3. The cells were starved for sulfate for 1 h, and then labeled for up to 75 min with [35S]sulfate in the presence of LaM4-3xTS nanobodies either in the absence (−) or presence of 500 nM rapamycin to inactivate GGA2-FKBP (+Rapa). The nanobodies were isolated by Ni/NTA beads and subjected to SDS-gel electrophoresis followed by immunoblot analysis (anti-His6) and autoradiography ([35S]). In parallel, aliquots of the cell lysates were immunoblotted for actin as a control for the amount of cells used. (C) Experiments as shown in panel B were quantified and presented as the percentage of the value in the absence of rapamycin after 75 min (mean and SD of three independent experiments). Without rapamycin is shown as black squares, with rapamycin as gray circles; uptake as open symbols, sulfation as filled symbols. (E) HeLa-GGA2ks cells stably expressing mCherry-CDMPR were siRNA-silenced for endogenous GGA1–3, followed by starvation for sulfate in the presence of LaM4-3xTS to preload mCherry-CDMPR in the surface/endosome/TGN pool. The cells were then labeled with [35S]sulfate for up to 75 min in the continued presence of LaM4-3xTS, without or with addition of 500 nM rapamycin after 15 min (arrow) to inactivate GGA2 (+Rapa). (B) Nanobody analysis was performed as above (B). (F) Experiments as shown in panel E were quantified and presented as the percentage of the value in the absence of rapamycin after 75 min (mean and SD of three independent experiments). Without rapamycin is shown as black squares, with rapamycin as gray circles; uptake as open symbols, sulfation as filled symbols.