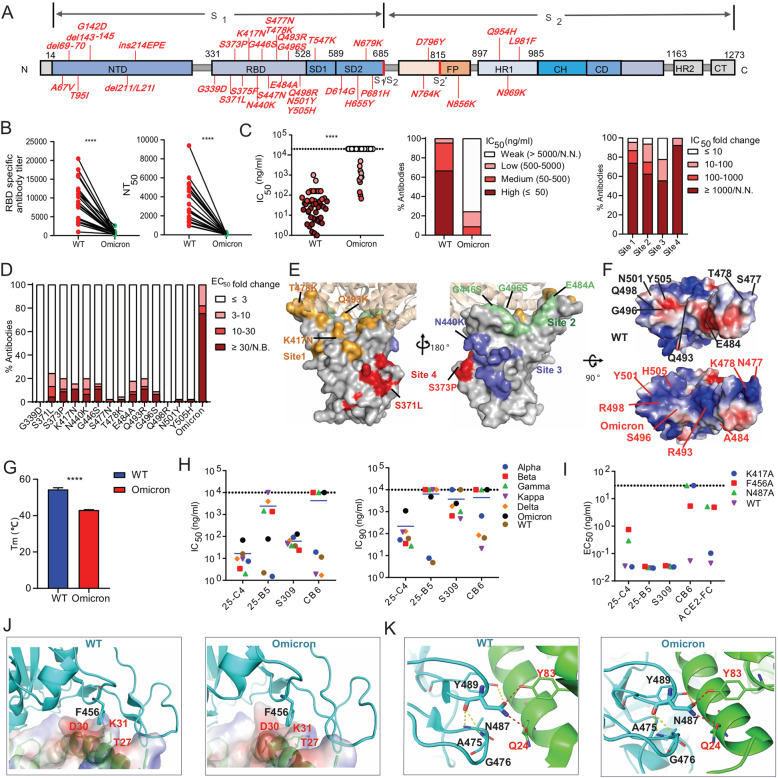
Fig. 1.
Immune evasion analysis of the Omicron variant and identification of a cross-variant neutralizing antibody targeting conserved RBD amino acids. A The key spike mutations present in the Omicron variant (B.1.1.529) are highlighted. B The anti-RBD antibody titer and neutralizing activity of the plasma from COVID-19 convalescent individuals (n = 21) against Wuhan-Hu-1 (wild-type, WT) and the Omicron variant. RBD-specific IgG antibody (Ab) titers were calculated as the endpoint dilution that remained positively detectable for the wild-type and Omicron RBD. Fifty percent neutralizing antibody titers (NT50) were calculated against wild-type and Omicron pseudoviruses. C Neutralization capability of Omicron pseudovirus by a panel of 45 RBD-specific mAbs. The left panel shows the IC50 values of the tested antibodies against wild-type and Omicron pseudoviruses. The middle panel shows the percentage of antibodies with the indicated neutralization activities. The right panel shows the percentage of different degrees of IC50 fold change for antibodies targeting four antigenic sites. The IC50 fold change is the ratio of the IC50 of antibodies against Omicron compared with wild-type pseudovirus. D The percentage of different degrees of antibody binding EC50 fold change of the Omicron RBD or single-amino acid substitutions present in Omicron. The EC50 fold change was calculated as the EC50 of the mutant RBD /the EC50 of the wild-type RBD. E The key immune escape substitutions found in Omicron are highlighted at four distinct antigenic sites in the RBD region. The color-coding scheme: site 1 (orange), site 2 (green), site 3 (slate blue), site 4 (red); ACE2 (wheat). Left and right are shown from different angles. F Electrostatic potential surface representation of the RBD from wild-type and Omicron. Electrostatic surface potentials are colored red and blue for negative and positive charges, respectively; white represents neutral residues. The wild-type positions are shown in black font and Omicron in red. G The melting temperature (Tm) of the recombinant RBD of wild-type and Omicron measured by nano differential scanning fluorimetry (nanoDSF). H IC50 and IC90 values of four RBD-specific NAbs against different SARS-CoV-2 VOC pseudoviruses. I Binding EC50 values of the tested NAbs or ACE2 with the F456A, N487A and K417A RBD mutants. J Structural interaction between F456 on RBD from wild-type and omicron with ACE2. The binding surface of ACE2 is shown by electrostatic surface representations. K Structural interaction between N487 on the RBD of wild-type and omicron with ACE2. Yellow dashed lines, intramolecular hydrogen bond within the RBD; red dashed lines, polar interactions between the RBD and ACE2. All experimentally obtained data represent one of two independent experiments. Nonneutralizing IC50 values were set to over 10 μg/ml, and nonbinding EC50 values were set to over 5 μg/ml. The structures of RBD-ACE2 complexes were obtained from PDB (6m0j for wild-type [12] and 7 WBP for Omicron [10]). P values shown in the figures were determined using paired two-tailed Student’s t tests (****P < 0.0001)