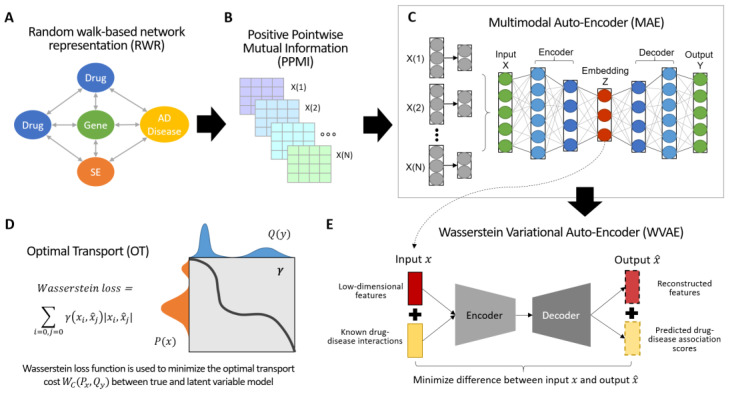
Figure 1.
Overview of DOTA. (A) Drug networks (drug–drug, drug–gene, drug–side-effects, drug–disease, and five other drug–drug similarities) are first converted into high-quality vector representation with a random walk-based procedure. (B) Next, the associations of the factorized co-occurrence matrixes are represented as PPMI matrixes. (C) The PPMI matrixes are then fused together into a low-dimensional feature representation using an unsupervised multimodal auto-encoder. (D) The optimal transport problem used in the second autoencoder part of DOTA. A Wasserstein loss function is used to minimize the optimal transport cost between the input and output. (E) The drug information from the embedding layer of MAE is extracted and used as drug features to predict new drug-disease associations. A WVAE is used to encode and decode the drug-associations.