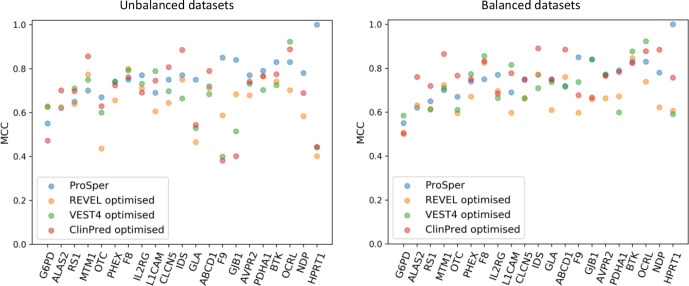
Figure 5.
A comparison of the Matthews correlation coefficient (MCC) values for ProSper (protein-specific variant interpreter) with the optimised MCC values for VEST4, REVEL and ClinPred using all of the data sets (on the left) and using balanced data sets (on the right). Optimised MCC values were generated using gene-specific or protein-specific pathogenicity thresholds. The data set for each gene was balanced using undersampling, that is, using a random subset from the majority class to match the number of variants in the minority class. The gene-specific threshold was identified using 80% of all the predictions from each tool through repeated (n=10) fivefold cross-validation with random subsampling. The optimised MCC value was generated using the rest (20%) of the predictions from each tool at the threshold identified for each gene. VEST4 predictions were unavailable for ALAS2 and NDP variants in the respective transcripts of interest.